Integration of transcriptome, proteome and metabolism data reveals the alkaloids biosynthesis in Macleaya cordata and Macleaya microcarpa
- PMID: 23326424
- PMCID: PMC3541140
- DOI: 10.1371/journal.pone.0053409
Integration of transcriptome, proteome and metabolism data reveals the alkaloids biosynthesis in Macleaya cordata and Macleaya microcarpa
Erratum in
- PLoS One. 2013;8(9). doi:10.1371/annotation/6848d2aa-d15f-4632-9074-727b25958da3
Abstract
Background: The Macleaya spp., including Macleaya cordata and Macleaya microcarpa, are traditional anti-virus, inflammation eliminating, and insecticide herb medicines for their isoquinoline alkaloids. They are also known as the basis of the popular natural animal food addictive in Europe. However, few studies especially at genomics level were conducted on them. Hence, we performed the Macleaya spp. transcriptome and integrated it with iTRAQ proteome analysis in order to identify potential genes involved in alkaloids biosynthesis.
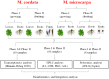
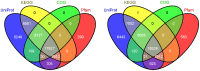
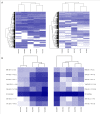
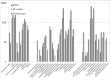
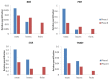
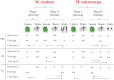
Methodology and principal findings: We elaborately designed the transcriptome, proteome and metabolism profiling for 10 samples of both species to explore their alkaloids biosynthesis. From the transcriptome data, we obtained 69367 and 78255 unigenes for M. cordata and M. microcarpa, in which about two thirds of them were similar to sequences in public databases. By metabolism profiling, reverse patterns for alkaloids sanguinarine, chelerythrine, protopine, and allocryptopine were observed in different organs of two species. We characterized the expressions of enzymes in alkaloid biosynthesis pathways. We also identified more than 1000 proteins from iTRAQ proteome data. Our results strongly suggest that the root maybe the organ for major alkaloids biosynthesis of Macleaya spp. Except for biosynthesis, the alkaloids storage and transport were also important for their accumulation. The ultrastructure of laticifers by SEM helps us to prove the alkaloids maybe accumulated in the mature roots.
Conclusions/significance: To our knowledge this is the first study to elucidate the genetic makeup of Macleaya spp. This work provides clues to the identification of the potential modulate genes involved in alkaloids biosynthesis in Macleaya spp., and sheds light on researches for non-model medicinal plants by integrating different high-throughput technologies.
Conflict of interest statement
Figures


References
-
- Psotova J, Zdarilova A, Anzenbacherova E, Kosina P, Svobodova A, et al. (2006) Safety assessment of sanguiritrin, alkaloid fraction of Macleaya cordata, in rats. Veterinarni Medicina 51: 145–155.
-
- Franz C, Bauer R, Carle R, Tedesco D, Tubaro A, et al... (2005) Study on the assessment of plants/herb extracts and their naturally or synthetically produced components as “additives” for use in animal production.: CFT/EFSA/FEEDAP/2005/01.
-
- Stiborova M, Vostalova J, Zdarilova A, Ulrichova J, Hudecek J, et al. (2008) Macleaya cordata extract and Sangrovit genotoxicity. Assessment in vivo. Biomed Pap Med Fac Univ Palacky Olomouc Czech Repub 152: 35–39. - PubMed
-
- Juskiewicz J, Gruzauskas R, Zdunczyk Z, Semaskaite A, Jankowski J, et al. (2011) Effects of dietary addition of Macleaya cordata alkaloid extract on growth performance, caecal indices and breast meat fatty acids profile in male broilers. J Anim Physiol Anim Nutr (Berl) 95: 171–178. - PubMed
-
- Ye F, Feng F, Liu W (2009) [Alkaloids from Macleaya cordata]. Zhongguo Zhong Yao Za Zhi 34: 1683–1686. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

