Intestinal T-cell responses in celiac disease - impact of celiac disease associated bacteria
- PMID: 23326425
- PMCID: PMC3541273
- DOI: 10.1371/journal.pone.0053414
Intestinal T-cell responses in celiac disease - impact of celiac disease associated bacteria
Abstract
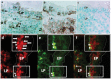
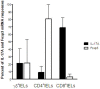
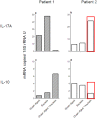
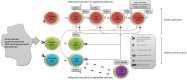
A hallmark of active celiac disease (CD), an inflammatory small-bowel enteropathy caused by permanent intolerance to gluten, is cytokine production by intestinal T lymphocytes. Prerequisites for contracting CD are that the individual carries the MHC class II alleles HLA-DQ2 and/or HLA-DQ8 and is exposed to gluten in the diet. Dysbiosis in the resident microbiota has been suggested to be another risk factor for CD. In fact, rod shaped bacteria adhering to the small intestinal mucosa were frequently seen in patients with CD during the "Swedish CD epidemic" and bacterial candidates could later be isolated from patients born during the epidemic suggesting long-lasting changes in the gut microbiota. Interleukin-17A (IL-17A) plays a role in both inflammation and anti-bacterial responses. In active CD IL-17A was produced by both CD8(+) T cells (Tc17) and CD4(+) T cells (Th17), with intraepithelial Tc17 cells being the dominant producers. Gluten peptides as well as CD associated bacteria induced IL-17A responses in ex vivo challenged biopsies from patients with inactive CD. The IL-17A response was suppressed in patients born during the epidemic when a mixture of CD associated bacteria was added to gluten, while the reverse was the case in patients born after the epidemic. Under these conditions Th17 cells were the dominant producers. Thus Tc17 and Th17 responses to gluten and bacteria seem to pave the way for the chronic disease with interferon-γ-production by intraepithelial Tc1 cells and lamina propria Th1 cells. The CD associated bacteria and the dysbiosis they might cause in the resident microbiota may be a risk factor for CD either by directly influencing the immune responses in the mucosa or by enhancing inflammatory responses to gluten.
Conflict of interest statement
Figures


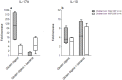

References
-
- Sollid LM (2002) Coeliac disease: dissecting a complex inflammatory disorder. Nat Rev Immunol 2: 647–655. - PubMed
-
- Tack GJ, Verbeek WH, Schreurs MW, Mulder CJ (2010) The spectrum of celiac disease: epidemiology, clinical aspects and treatment. Nat Rev Gastroenterol Hepatol 7: 204–213. - PubMed
-
- Di Sabatino A, Corazza GR (2009) Coeliac disease. Lancet 373: 1480–1493. - PubMed
-
- Forsberg G, Hernell O, Melgar S, Israelsson A, Hammarström S, et al. (2002) Paradoxical coexpression of proinflammatory and down-regulatory cytokines in intestinal T cells in childhood celiac disease. Gastroenterology 123: 667–678. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

