Insights into the genetic architecture of early stage age-related macular degeneration: a genome-wide association study meta-analysis
- PMID: 23326517
- PMCID: PMC3543264
- DOI: 10.1371/journal.pone.0053830
Insights into the genetic architecture of early stage age-related macular degeneration: a genome-wide association study meta-analysis
Abstract
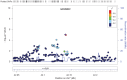
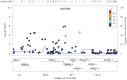
Genetic factors explain a majority of risk variance for age-related macular degeneration (AMD). While genome-wide association studies (GWAS) for late AMD implicate genes in complement, inflammatory and lipid pathways, the genetic architecture of early AMD has been relatively under studied. We conducted a GWAS meta-analysis of early AMD, including 4,089 individuals with prevalent signs of early AMD (soft drusen and/or retinal pigment epithelial changes) and 20,453 individuals without these signs. For various published late AMD risk loci, we also compared effect sizes between early and late AMD using an additional 484 individuals with prevalent late AMD. GWAS meta-analysis confirmed previously reported association of variants at the complement factor H (CFH) (peak P = 1.5×10(-31)) and age-related maculopathy susceptibility 2 (ARMS2) (P = 4.3×10(-24)) loci, and suggested Apolipoprotein E (ApoE) polymorphisms (rs2075650; P = 1.1×10(-6)) associated with early AMD. Other possible loci that did not reach GWAS significance included variants in the zinc finger protein gene GLI3 (rs2049622; P = 8.9×10(-6)) and upstream of GLI2 (rs6721654; P = 6.5×10(-6)), encoding retinal Sonic hedgehog signalling regulators, and in the tyrosinase (TYR) gene (rs621313; P = 3.5×10(-6)), involved in melanin biosynthesis. For a range of published, late AMD risk loci, estimated effect sizes were significantly lower for early than late AMD. This study confirms the involvement of multiple established AMD risk variants in early AMD, but suggests weaker genetic effects on the risk of early AMD relative to late AMD. Several biological processes were suggested to be potentially specific for early AMD, including pathways regulating RPE cell melanin content and signalling pathways potentially involved in retinal regeneration, generating hypotheses for further investigation.
Conflict of interest statement
Figures

References
-
- Lim LS, Mitchell P, Seddon JM, Holz FG, Wong TY (2012) Age-related macular degeneration. Lancet 379: 1728–1738. - PubMed
-
- Sallo FB, Peto T, Leung I, Xing W, Bunce C, et al. (2009) The International Classification system and the progression of age-related macular degeneration. Curr Eye Res 34: 238–240. - PubMed
-
- Kinnunen K, Petrovski G, Moe MC, Berta A, Kaarniranta K (2011) Molecular mechanisms of retinal pigment epithelium damage and development of age-related macular degeneration. Acta Ophthalmol. - PubMed
-
- Abdelsalam A, Del Priore L, Zarbin MA (1999) Drusen in age-related macular degeneration: pathogenesis, natural course, and laser photocoagulation-induced regression. Surv Ophthalmol 44: 1–29. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- HHSN268201100009I/HL/NHLBI NIH HHS/United States
- UL1TR000124/TR/NCATS NIH HHS/United States
- N01 AG012100/AG/NIA NIH HHS/United States
- N01-HC-85082/HC/NHLBI NIH HHS/United States
- R01 AG020098/AG/NIA NIH HHS/United States
- HL080295/HL/NHLBI NIH HHS/United States
- N01-HC-85084/HC/NHLBI NIH HHS/United States
- R01HL086694/HL/NHLBI NIH HHS/United States
- HHSN268201100012C/HL/NHLBI NIH HHS/United States
- UL1RR025005/RR/NCRR NIH HHS/United States
- HHSN268201100010C/HL/NHLBI NIH HHS/United States
- N01-HC-85085/HC/NHLBI NIH HHS/United States
- R01HL59367/HL/NHLBI NIH HHS/United States
- UL1 RR025005/RR/NCRR NIH HHS/United States
- R01 AG015928/AG/NIA NIH HHS/United States
- ZIA EY000426/ImNIH/Intramural NIH HHS/United States
- HHSN268201100008C/HL/NHLBI NIH HHS/United States
- U01 HL080295/HL/NHLBI NIH HHS/United States
- HHSN268201100005G/HL/NHLBI NIH HHS/United States
- Z99 EY999999/ImNIH/Intramural NIH HHS/United States
- Z01 EY000426/ImNIH/Intramural NIH HHS/United States
- HHSN268201100008I/HL/NHLBI NIH HHS/United States
- N01-HC-85081/HC/NHLBI NIH HHS/United States
- ZIA EY000401/ImNIH/Intramural NIH HHS/United States
- R01 HL059367/HL/NHLBI NIH HHS/United States
- HHSN268201100007C/HL/NHLBI NIH HHS/United States
- HL105756/HL/NHLBI NIH HHS/United States
- N01 HC015103/HC/NHLBI NIH HHS/United States
- N01 HC085086/HL/NHLBI NIH HHS/United States
- ZIAEY000401/PHS HHS/United States
- AG-15928/AG/NIA NIH HHS/United States
- R56 AG020098/AG/NIA NIH HHS/United States
- 4584/CRUK_/Cancer Research UK/United Kingdom
- HHSN268201100011I/HL/NHLBI NIH HHS/United States
- HHSN268201100011C/HL/NHLBI NIH HHS/United States
- R01 HL086694/HL/NHLBI NIH HHS/United States
- 085475/B/08/Z/WT_/Wellcome Trust/United Kingdom
- R01 HL087652/HL/NHLBI NIH HHS/United States
- AG-20098/AG/NIA NIH HHS/United States
- ZIA AG007380/ImNIH/Intramural NIH HHS/United States
- HL087652/HL/NHLBI NIH HHS/United States
- UL1 TR000124/TR/NCATS NIH HHS/United States
- HHSN268200625226C/PHS HHS/United States
- U01 HG004402/HG/NHGRI NIH HHS/United States
- N01-HC-85086/HC/NHLBI NIH HHS/United States
- ZIAAG007380/PHS HHS/United States
- R01 HL105756/HL/NHLBI NIH HHS/United States
- U01HG004402/HG/NHGRI NIH HHS/United States
- AG-027058/AG/NIA NIH HHS/United States
- P30 DK063491/DK/NIDDK NIH HHS/United States
- HHSN268201100006C/HL/NHLBI NIH HHS/United States
- N01 HC-55222/HC/NHLBI NIH HHS/United States
- HHSN268201200036C/HL/NHLBI NIH HHS/United States
- R01HL087641/HL/NHLBI NIH HHS/United States
- N01 HC055222/HL/NHLBI NIH HHS/United States
- HHSN268201100005I/HL/NHLBI NIH HHS/United States
- N01 HC085079/HL/NHLBI NIH HHS/United States
- N01-HC-85083/HC/NHLBI NIH HHS/United States
- N01-HC-85080/HC/NHLBI NIH HHS/United States
- 090532/WT_/Wellcome Trust/United Kingdom
- N01-HC-75150/HC/NHLBI NIH HHS/United States
- R01 HL080295/HL/NHLBI NIH HHS/United States
- Z01 EY000401/ImNIH/Intramural NIH HHS/United States
- HHSN268201100009C/HL/NHLBI NIH HHS/United States
- N01-AG-12100/AG/NIA NIH HHS/United States
- HHSN268201100005C/HL/NHLBI NIH HHS/United States
- DK063491/DK/NIDDK NIH HHS/United States
- N01-HC-45133/HC/NHLBI NIH HHS/United States
- HHSN268201100007I/HL/NHLBI NIH HHS/United States
- N01-HC-85079/HC/NHLBI NIH HHS/United States
- N01-HC-85239/HC/NHLBI NIH HHS/United States
- AG-023629/AG/NIA NIH HHS/United States
- N01 HC075150/HL/NHLBI NIH HHS/United States
- R01 HL087641/HL/NHLBI NIH HHS/United States
- R01 AG023629/AG/NIA NIH HHS/United States
- 085475/08/Z/WT_/Wellcome Trust/United Kingdom
- R01 AG027058/AG/NIA NIH HHS/United States
- N01 HC045133/HC/NHLBI NIH HHS/United States
- N01 HC035129/HC/NHLBI NIH HHS/United States
- R56 AG023629/AG/NIA NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

