de novo design and synthesis of Candida antarctica lipase B gene and α-factor leads to high-level expression in Pichia pastoris
- PMID: 23326544
- PMCID: PMC3542265
- DOI: 10.1371/journal.pone.0053939
de novo design and synthesis of Candida antarctica lipase B gene and α-factor leads to high-level expression in Pichia pastoris
Abstract
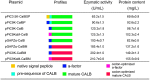
Candida antarctica lipase B (CALB) is one of the most widely used and studied enzymes in the world. In order to achieve the high-level expression of CALB in Pichia, we optimized the codons of CALB gene and α-factor by using a de novo design and synthesis strategy. Through comparative analysis of a series of recombinants with different expression components, we found that the methanol-inducible expression recombinant carrying the codon-optimized α-factor and mature CALB gene (pPIC9KαM-CalBM) has the highest lipase production capacity. After fermentation parameters optimization, the lipase activity and protein content of the recombinant pPIC9KαM-CalBM reached 6,100 U/mL and 3.0 g/L, respectively, in a 5-L fermentor. We believe this strategy could be of special interest due to its capacity to improve the expression level of target gene, and the Pichia transformants carrying the codon-optimized gene had great potential for the industrial-scale production of CALB lipase.
Conflict of interest statement
Figures



References
-
- Patkar SA, BjoÅNrking F, Zundel M, Schulein M, Svendsen A, et al. (1993) Purification of two lipases from Candida antarcticaand their inhibition by various inhibitors. Indian J Chem Sect B 32: 76–80.
-
- Jaeger KE, Dijkstra BW, Reetz MT (1999) Bacterial biocatalysis: molecular biology, three-dimensional structures, and biotechnological applications of lipases. Annu Rev Microbiol 53: 315–51. - PubMed
-
- Yu Y, Lutz S (2010) Improved triglyceride transesterification by circular permuted Candida antarctica lipase B. Biotechnol Bioeng. 105: 44–50. - PubMed
-
- Ko MJ, Park HJ, Hong SY, Yoo YJ (2012) Continuous biodiesel production using in situ glycerol separation by membrane bioreactor system. Bioprocess Biosyst Eng 35: 69–75. - PubMed
-
- Tanino T, Aoki T, Chung WY, Watanabe Y, Ogino C, et al. (2009) Improvement of a Candida antarctica lipase B-displaying yeast whole-cell biocatalyst and its application to the polyester synthesis reaction. Appl Microbiol Biotechnol 82: 59–66. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

