Detection of Leishmania RNA virus in Leishmania parasites
- PMID: 23326619
- PMCID: PMC3542153
- DOI: 10.1371/journal.pntd.0002006
Detection of Leishmania RNA virus in Leishmania parasites
Abstract
Background: Patients suffering from cutaneous leishmaniasis (CL) caused by New World Leishmania (Viannia) species are at high risk of developing mucosal (ML) or disseminated cutaneous leishmaniasis (DCL). After the formation of a primary skin lesion at the site of the bite by a Leishmania-infected sand fly, the infection can disseminate to form secondary lesions. This metastatic phenotype causes significant morbidity and is often associated with a hyper-inflammatory immune response leading to the destruction of nasopharyngeal tissues in ML, and appearance of nodules or numerous ulcerated skin lesions in DCL. Recently, we connected this aggressive phenotype to the presence of Leishmania RNA virus (LRV) in strains of L. guyanensis, showing that LRV is responsible for elevated parasitaemia, destructive hyper-inflammation and an overall exacerbation of the disease. Further studies of this relationship and the distribution of LRVs in other Leishmania strains and species would benefit from improved methods of viral detection and quantitation, especially ones not dependent on prior knowledge of the viral sequence as LRVs show significant evolutionary divergence.
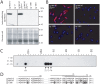
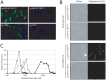
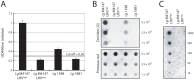
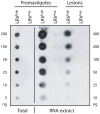
Methodology/principal findings: This study reports various techniques, among which, the use of an anti-dsRNA monoclonal antibody (J2) stands out for its specific and quantitative recognition of dsRNA in a sequence-independent fashion. Applications of J2 include immunofluorescence, ELISA and dot blot: techniques complementing an arsenal of other detection tools, such as nucleic acid purification and quantitative real-time-PCR. We evaluate each method as well as demonstrate a successful LRV detection by the J2 antibody in several parasite strains, a freshly isolated patient sample and lesion biopsies of infected mice.
Conclusions/significance: We propose that refinements of these methods could be transferred to the field for use as a diagnostic tool in detecting the presence of LRV, and potentially assessing the LRV-related risk of complications in cutaneous leishmaniasis.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




Similar articles
-
Leishmania aethiopica field isolates bearing an endosymbiontic dsRNA virus induce pro-inflammatory cytokine response.PLoS Negl Trop Dis. 2014 Apr 24;8(4):e2836. doi: 10.1371/journal.pntd.0002836. eCollection 2014 Apr. PLoS Negl Trop Dis. 2014. PMID: 24762979 Free PMC article.
-
Leishmania RNA virus: when the host pays the toll.Front Cell Infect Microbiol. 2012 Jul 12;2:99. doi: 10.3389/fcimb.2012.00099. eCollection 2012. Front Cell Infect Microbiol. 2012. PMID: 22919688 Free PMC article. Review.
-
MyD88 and TLR9 dependent immune responses mediate resistance to Leishmania guyanensis infections, irrespective of Leishmania RNA virus burden.PLoS One. 2014 May 6;9(5):e96766. doi: 10.1371/journal.pone.0096766. eCollection 2014. PLoS One. 2014. PMID: 24801628 Free PMC article.
-
Prevalence of Leishmania RNA virus in Leishmania parasites in patients with tegumentary leishmaniasis: A systematic review and meta-analysis.PLoS Negl Trop Dis. 2022 Jun 8;16(6):e0010427. doi: 10.1371/journal.pntd.0010427. eCollection 2022 Jun. PLoS Negl Trop Dis. 2022. PMID: 35675332 Free PMC article.
-
Detection of Leishmania RNA Virus in Clinical Samples from Cutaneous Leishmaniasis Patients Varies according to the Type of Sample.Am J Trop Med Hyg. 2021 Jan;104(1):233-239. doi: 10.4269/ajtmh.20-0073. Am J Trop Med Hyg. 2021. PMID: 33146111 Free PMC article.
Cited by
-
Leishmania guyanensis M4147 as a new LRV1-bearing model parasite: Phosphatidate phosphatase 2-like protein controls cell cycle progression and intracellular lipid content.PLoS Negl Trop Dis. 2022 Jun 24;16(6):e0010510. doi: 10.1371/journal.pntd.0010510. eCollection 2022 Jun. PLoS Negl Trop Dis. 2022. PMID: 35749562 Free PMC article.
-
Correlation between presence of Leishmania RNA virus 1 and clinical characteristics of nasal mucosal leishmaniosis.Braz J Otorhinolaryngol. 2015 Sep-Oct;81(5):533-40. doi: 10.1016/j.bjorl.2015.07.014. Epub 2015 Jul 22. Braz J Otorhinolaryngol. 2015. PMID: 26277588 Free PMC article.
-
Identification of divergent Leishmania (Viannia) braziliensis ecotypes derived from a geographically restricted area through whole genome analysis.PLoS Negl Trop Dis. 2019 Jun 6;13(6):e0007382. doi: 10.1371/journal.pntd.0007382. eCollection 2019 Jun. PLoS Negl Trop Dis. 2019. PMID: 31170148 Free PMC article.
-
New insights into the genetic diversity of Leishmania RNA Virus 1 and its species-specific relationship with Leishmania parasites.PLoS One. 2018 Jun 18;13(6):e0198727. doi: 10.1371/journal.pone.0198727. eCollection 2018. PLoS One. 2018. PMID: 29912912 Free PMC article.
-
Transcriptomics analysis highlights potential ways in human pathogenesis in Leishmania braziliensis infected with the viral endosymbiont LRV1.PLoS Negl Trop Dis. 2024 May 14;18(5):e0012126. doi: 10.1371/journal.pntd.0012126. eCollection 2024 May. PLoS Negl Trop Dis. 2024. PMID: 38743668 Free PMC article.
References
-
- den Boer M, Argaw D, Jannin J, Alvar J (2011) Leishmaniasis impact and treatment access. Clin Microbiol Infect 17: 1471–1477. - PubMed
-
- Banuls AL, Bastien P, Pomares C, Arevalo J, Fisa R, et al. (2011) Clinical pleiomorphism in human leishmaniases, with special mention of asymptomatic infection. Clin Microbiol Infect 17: 1451–1461. - PubMed
-
- Santrich C, Segura I, Arias AL, Saravia NG (1990) Mucosal disease caused by Leishmania braziliensis guyanensis . Am J Trop Med Hyg 42: 51–55. - PubMed
-
- Weigle K, Saravia NG (1996) Natural history, clinical evolution, and the host-parasite interaction in New World cutaneous Leishmaniasis. Clin Dermatol 14: 433–450. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

