Gene gymnastics: Synthetic biology for baculovirus expression vector system engineering
- PMID: 23328086
- PMCID: PMC3813527
- DOI: 10.4161/bioe.22966
Gene gymnastics: Synthetic biology for baculovirus expression vector system engineering
Abstract
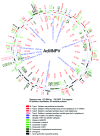
Most essential activities in eukaryotic cells are catalyzed by large multiprotein assemblies containing up to ten or more interlocking subunits. The vast majority of these protein complexes are not easily accessible for high resolution studies aimed at unlocking their mechanisms, due to their low cellular abundance and high heterogeneity. Recombinant overproduction can resolve this bottleneck and baculovirus expression vector systems (BEVS) have emerged as particularly powerful tools for the provision of eukaryotic multiprotein complexes in high quality and quantity. Recently, synthetic biology approaches have begun to make their mark in improving existing BEVS reagents by de novo design of streamlined transfer plasmids and by engineering the baculovirus genome. Here we present OmniBac, comprising new custom designed reagents that further facilitate the integration of heterologous genes into the baculovirus genome for multiprotein expression. Based on comparative genome analysis and data mining, we herein present a blueprint to custom design and engineer the entire baculovirus genome for optimized production properties using a bottom-up synthetic biology approach.
Keywords: BEVS; MultiBac; OmniBac; baculovirus; comparative genome analysis; multiprotein complexes; recombinant protein production.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
