Autophagy and Crohn's disease
- PMID: 23328432
- PMCID: PMC6741541
- DOI: 10.1159/000345129
Autophagy and Crohn's disease
Abstract
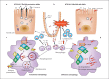
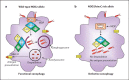
Advances in genetics have shed light on the molecular basis of Crohn's disease (CD) predisposition and pathogenesis, via linkage disequilibrium analysis to genome-wide association studies. The discovery of genetic variants of NOD2, an intracellular pathogen molecular sensor, as risk factors for CD has paved the way for further research on innate immunity in this disease. Remarkably, polymorphisms in autophagy genes, such as ATG16L1 and IRGM, have been identified, allowing the pivotal role of autophagy in innate immunity to be uncovered. In this review, we summarize recent studies on the CD-associated NOD2, ATG16L1 and IRGM risk variants and their contribution to the autophagy functions that have most influenced our understanding of CD pathophysiology.
Copyright © 2013 S. Karger AG, Basel.
Figures
References
-
- Blumberg R, Cho J, Lewis J, Wu G. Inflammatory bowel disease: an update on the fundamental biology and clinical management. Gastroenterology. 2011;140:1701–1703. - PubMed
-
- Chassaing B, Darfeuille-Michaud A. The commensal microbiota and enteropathogens in the pathogenesis of inflammatory bowel diseases. Gastroenterology. 2011;140:1720–1728. - PubMed
-
- Rosenstiel P, Sina C, Franke A, Schreiber S. Towards a molecular risk map - recent advances on the etiology of inflammatory bowel disease. Semin Immunol. 2009;21:334–345. - PubMed
-
- Hugot JP, Chamaillard M, Zouali H, Lesage S, Cezard JP, Belaiche J, Almer S, Tysk C, O'Morain CA, Gassull M, Binder V, Finkel Y, Cortot A, Modigliani R, Laurent-Puig P, Gower-Rousseau C, Macry J, Colombel JF, Sahbatou M, Thomas G. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn's disease. Nature. 2001;411:599–603. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

