Phenotypic profiling of DPYD variations relevant to 5-fluorouracil sensitivity using real-time cellular analysis and in vitro measurement of enzyme activity
- PMID: 23328581
- PMCID: PMC3602211
- DOI: 10.1158/0008-5472.CAN-12-3858
Phenotypic profiling of DPYD variations relevant to 5-fluorouracil sensitivity using real-time cellular analysis and in vitro measurement of enzyme activity
Abstract
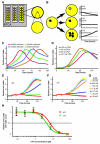
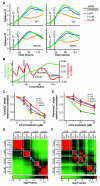
In the 45 years since its development, the pyrimidine analog 5-fluorouracil (5-FU) has become an integral component of many cancer treatments, most notably for the management of colorectal cancer. An appreciable fraction of patients who receive 5-FU suffer severe adverse toxicities, which in extreme cases may result in death. Dihydropyrimidine dehydrogenase (DPD, encoded by DPYD) rapidly degrades 85% of administered 5-FU, and as such, limits the amount of drug available for conversion into active metabolites. Clinical studies have suggested that genetic variations in DPYD increase the risk for 5-FU toxicity, however, there is not a clear consensus about which variations are relevant predictors. In the present study, DPYD variants were expressed in mammalian cells, and the enzymatic activity of expressed protein was determined relative to wild-type (WT). Relative sensitivity to 5-FU for cells expressing DPYD variations was also measured. The DPYD*2A variant (exon 14 deletion caused by IVS14+1G>A) was confirmed to be catalytically inactive. Compared with WT, two variants, S534N and C29R, showed significantly higher enzymatic activity. Cells expressing S534N were more resistant to 5-FU-mediated toxicity compared with cells expressing WT DPYD. These findings support the hypothesis that selected DPYD alleles are protective against severe 5-FU toxicity, and, as a consequence, may decrease the effectiveness of 5-FU an antitumor drug in carriers. In addition, this study shows a method that may be useful for phenotyping other genetic variations in pharmacologically relevant pathways.
Figures

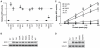
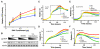
References
-
- Rich TA, Shepard RC, Mosley ST. Four decades of continuing innovation with fluorouracil: current and future approaches to fluorouracil chemoradiation therapy. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2004;22:2214–32. - PubMed
-
- MacMillan WE, Wolberg WH, Welling PG. Pharmacokinetics of fluorouracil in humans. Cancer Res. 1978;38:3479–82. - PubMed
-
- Heggie GD, Sommadossi JP, Cross DS, Huster WJ, Diasio RB. Clinical pharmacokinetics of 5-fluorouracil and its metabolites in plasma, urine, and bile. Cancer Res. 1987;47:2203–6. - PubMed
-
- Longley DB, Harkin DP, Johnston PG. 5-fluorouracil: mechanisms of action and clinical strategies. Nat Rev Cancer. 2003;3:330–8. - PubMed
-
- Harris BE, Song R, Soong SJ, Diasio RB. Relationship between dihydropyrimidine dehydrogenase activity and plasma 5-fluorouracil levels with evidence for circadian variation of enzyme activity and plasma drug levels in cancer patients receiving 5-fluorouracil by protracted continuous infusion. Cancer Res. 1990;50:197–201. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

