Nucleosomal composition at the centromere: a numbers game
- PMID: 23328870
- PMCID: PMC3601254
- DOI: 10.1007/s10577-012-9335-7
Nucleosomal composition at the centromere: a numbers game
Abstract
The Centromere is a unique chromosomal locus where the kinetochore is formed to mediate faithful chromosome partitioning, thus maintaining ploidy during cell division. Centromere identity is inherited via an epigenetic mechanism involving a histone H3 variant, called centromere protein A (CENP-A) which replaces H3 in centromeric chromatin. In spite of extensive efforts in field of centromere biology during the past decade, controversy persists over the structural nature of the CENP-A-containing epigenetic mark, both at nucleosomal and chromatin levels. Here, we review recent findings and hypotheses regarding the structure of CENP-A-containing complexes.
Figures
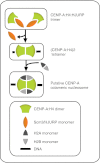

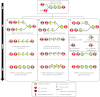
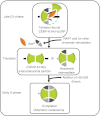
Similar articles
-
Basic properties of epigenetic systems: lessons from the centromere.Curr Opin Genet Dev. 2013 Apr;23(2):219-27. doi: 10.1016/j.gde.2012.11.002. Epub 2012 Dec 5. Curr Opin Genet Dev. 2013. PMID: 23219400 Review.
-
Human centromeric CENP-A chromatin is a homotypic, octameric nucleosome at all cell cycle points.J Cell Biol. 2017 Mar 6;216(3):607-621. doi: 10.1083/jcb.201608083. Epub 2017 Feb 24. J Cell Biol. 2017. PMID: 28235947 Free PMC article.
-
Octameric CENP-A nucleosomes are present at human centromeres throughout the cell cycle.Curr Biol. 2013 May 6;23(9):764-9. doi: 10.1016/j.cub.2013.03.037. Epub 2013 Apr 25. Curr Biol. 2013. PMID: 23623556
-
Preparation of Recombinant Centromeric Nucleosomes and Formation of Complexes with Nonhistone Centromere Proteins.Methods Enzymol. 2016;573:67-96. doi: 10.1016/bs.mie.2016.01.014. Epub 2016 Feb 23. Methods Enzymol. 2016. PMID: 27372749
-
Focus on the centre: the role of chromatin on the regulation of centromere identity and function.EMBO J. 2009 Aug 19;28(16):2337-48. doi: 10.1038/emboj.2009.174. Epub 2009 Jul 23. EMBO J. 2009. PMID: 19629040 Free PMC article. Review.
Cited by
-
Phase-plate cryo-EM structure of the Widom 601 CENP-A nucleosome core particle reveals differential flexibility of the DNA ends.Nucleic Acids Res. 2020 Jun 4;48(10):5735-5748. doi: 10.1093/nar/gkaa246. Nucleic Acids Res. 2020. PMID: 32313946 Free PMC article.
-
Bending the rules: widefield microscopy and the Abbe limit of resolution.J Cell Physiol. 2014 Feb;229(2):132-8. doi: 10.1002/jcp.24439. J Cell Physiol. 2014. PMID: 23893718 Free PMC article. Review.
-
Structure of centromere chromatin: from nucleosome to chromosomal architecture.Chromosoma. 2017 Aug;126(4):443-455. doi: 10.1007/s00412-016-0620-7. Epub 2016 Nov 17. Chromosoma. 2017. PMID: 27858158 Free PMC article. Review.
-
A Molecular View of Kinetochore Assembly and Function.Biology (Basel). 2017 Jan 24;6(1):5. doi: 10.3390/biology6010005. Biology (Basel). 2017. PMID: 28125021 Free PMC article. Review.
-
Discovering centromere proteins: from cold white hands to the A, B, C of CENPs.Nat Rev Mol Cell Biol. 2015 Jul;16(7):443-9. doi: 10.1038/nrm4001. Epub 2015 May 20. Nat Rev Mol Cell Biol. 2015. PMID: 25991376
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

