RNA interference in the nucleus: roles for small RNAs in transcription, epigenetics and beyond
- PMID: 23329111
- PMCID: PMC4205957
- DOI: 10.1038/nrg3355
RNA interference in the nucleus: roles for small RNAs in transcription, epigenetics and beyond
Abstract
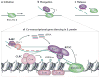
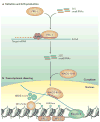
A growing number of functions are emerging for RNA interference (RNAi) in the nucleus, in addition to well-characterized roles in post-transcriptional gene silencing in the cytoplasm. Epigenetic modifications directed by small RNAs have been shown to cause transcriptional repression in plants, fungi and animals. Additionally, increasing evidence indicates that RNAi regulates transcription through interaction with transcriptional machinery. Nuclear small RNAs include small interfering RNAs (siRNAs) and PIWI-interacting RNAs (piRNAs) and are implicated in nuclear processes such as transposon regulation, heterochromatin formation, developmental gene regulation and genome stability.
Conflict of interest statement
The authors declare that they have no competing financial interests
Figures


References
-
- Aravin A, et al. A novel class of small RNAs bind to MILI protein in mouse testes. Nature. 2006;442:203–207. - PubMed
-
- Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol. 2009;10:126–139. - PubMed
-
- Colmenares SU, Buker SM, Bühler M, Dlakić M, Moazed D. Coupling of double-stranded RNA synthesis and siRNA generation in fission yeast RNAi. Mol Cell. 2007;27:449–461. - PubMed
-
- Pak J, Fire A. Distinct populations of primary and secondary effectors during RNAi in C. elegans. Science. 2007;315:241–244. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

