Y-chromosome evolution: emerging insights into processes of Y-chromosome degeneration
- PMID: 23329112
- PMCID: PMC4120474
- DOI: 10.1038/nrg3366
Y-chromosome evolution: emerging insights into processes of Y-chromosome degeneration
Abstract
The human Y chromosome is intriguing not only because it harbours the master-switch gene that determines gender but also because of its unusual evolutionary history. The Y chromosome evolved from an autosome, and its evolution has been characterized by massive gene decay. Recent whole-genome and transcriptome analyses of Y chromosomes in humans and other primates, in Drosophila species and in plants have shed light on the current gene content of the Y chromosome, its origins and its long-term fate. Furthermore, comparative analysis of young and old Y chromosomes has given further insights into the evolutionary and molecular forces triggering Y-chromosome degeneration and into the evolutionary destiny of the Y chromosome.
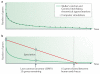
Figures


References
-
- Lahn BT, Page DC. Four evolutionary strata on the human X chromosome. Science. 1999;286:964–967. - PubMed
-
- Charlesworth B. The evolution of sex chromosomes. Science. 1990;251:1030–1033. - PubMed
-
- Bull JJ. Evolution of Sex Determining Mechanisms. Benjamin Cummings; Menlo Park, CA: 1983.
-
- Rice WR. Evolution of the Y sex chromosome in animals. BioScience. 1996;46:331–343.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

