Genetic and chemical characterization of ibuprofen degradation by Sphingomonas Ibu-2
- PMID: 23329679
- PMCID: PMC4083657
- DOI: 10.1099/mic.0.062273-0
Genetic and chemical characterization of ibuprofen degradation by Sphingomonas Ibu-2
Abstract
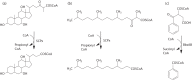
Sphingomonas Ibu-2 has the unusual ability to cleave the acid side chain from the pharmaceutical ibuprofen and related arylacetic acid derivatives to yield corresponding catechols under aerobic conditions via a previously uncharacterized mechanism. Screening a chromosomal library of Ibu-2 DNA in Escherichia coli EPI300 allowed us to identify one fosmid clone (pFOS3G7) that conferred the ability to metabolize ibuprofen to isobutylcatechol. Characterization of pFOS3G7 loss-of-function transposon mutants permitted identification of five ORFs, ipfABDEF, whose predicted amino acid sequences bore similarity to the large and small units of an aromatic dioxygenase (ipfAB), a sterol carrier protein X (SCPx) thiolase (ipfD), a domain of unknown function 35 (DUF35) protein (ipfE) and an aromatic CoA ligase (ipfF). Two additional ORFs, ipfH and ipfI, which encode putative ferredoxin reductase and ferredoxin components of an aromatic dioxygenase system, respectively, were also identified on pFOS3G7. Complementation of a markerless loss-of-function ipfD deletion mutant restored catechol production as did complementation of the ipfF Tn mutant. Expression of subcloned ipfABDEF alone in E. coli did not impart full metabolic activity unless coexpressed with ipfHI. CoA ligation followed by ring oxidation is common to phenylacetic acid pathways. However, the need for a putative SCPx thiolase (IpfD) and DUF35 protein (IpfE) in aerobic arylacetic acid degradation is unprecedented. This work provides preliminary insights into the mechanism behind this novel arylacetic acid-deacylating, catechol-generating activity.
Figures

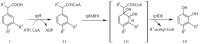
Similar articles
-
Formation of catechols via removal of acid side chains from ibuprofen and related aromatic acids.Appl Environ Microbiol. 2005 Oct;71(10):6121-5. doi: 10.1128/AEM.71.10.6121-6125.2005. Appl Environ Microbiol. 2005. PMID: 16204529 Free PMC article.
-
Patchwork assembly of nag-like nitroarene dioxygenase genes and the 3-chlorocatechol degradation cluster for evolution of the 2-chloronitrobenzene catabolism pathway in Pseudomonas stutzeri ZWLR2-1.Appl Environ Microbiol. 2011 Jul;77(13):4547-52. doi: 10.1128/AEM.02543-10. Epub 2011 May 20. Appl Environ Microbiol. 2011. PMID: 21602392 Free PMC article.
-
Tetralin-induced and ThnR-regulated aldehyde dehydrogenase and beta-oxidation genes in Sphingomonas macrogolitabida strain TFA.Appl Environ Microbiol. 2010 Jan;76(1):110-8. doi: 10.1128/AEM.01846-09. Epub 2009 Nov 6. Appl Environ Microbiol. 2010. PMID: 19897762 Free PMC article.
-
Pathway and evolutionary implications of diphenylamine biodegradation by Burkholderia sp. strain JS667.Appl Environ Microbiol. 2009 May;75(9):2694-704. doi: 10.1128/AEM.02198-08. Epub 2009 Feb 27. Appl Environ Microbiol. 2009. PMID: 19251893 Free PMC article.
-
The genes coding for the conversion of carbazole to catechol are flanked by IS6100 elements in Sphingomonas sp. strain XLDN2-5.PLoS One. 2010 Apr 2;5(4):e10018. doi: 10.1371/journal.pone.0010018. PLoS One. 2010. PMID: 20368802 Free PMC article.
Cited by
-
Organic micropollutants paracetamol and ibuprofen-toxicity, biodegradation, and genetic background of their utilization by bacteria.Environ Sci Pollut Res Int. 2018 Aug;25(22):21498-21524. doi: 10.1007/s11356-018-2517-x. Epub 2018 Jun 19. Environ Sci Pollut Res Int. 2018. PMID: 29923050 Free PMC article. Review.
-
Microbial degradation of contaminants of emerging concern: metabolic, genetic and omics insights for enhanced bioremediation.Front Bioeng Biotechnol. 2024 Sep 19;12:1470522. doi: 10.3389/fbioe.2024.1470522. eCollection 2024. Front Bioeng Biotechnol. 2024. PMID: 39364263 Free PMC article. Review.
-
New bacterial strains for ibuprofen biodegradation: Drug removal, transformation, and potential catabolic genes.Environ Microbiol Rep. 2024 Aug;16(4):e13320. doi: 10.1111/1758-2229.13320. Environ Microbiol Rep. 2024. PMID: 39187308 Free PMC article.
-
Ibuprofen Degradation and Associated Bacterial Communities in Hyporheic Zone Sediments.Microorganisms. 2020 Aug 16;8(8):1245. doi: 10.3390/microorganisms8081245. Microorganisms. 2020. PMID: 32824323 Free PMC article.
-
Ibuprofen: Toxicology and Biodegradation of an Emerging Contaminant.Molecules. 2023 Feb 23;28(5):2097. doi: 10.3390/molecules28052097. Molecules. 2023. PMID: 36903343 Free PMC article. Review.
References
-
- Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. (1990). Basic local alignment search tool. J Mol Biol 215, 403–410. - PubMed
-
- Aziz R., Saad R., Rizkallah M. (2011). PharmacoMicrobiomics or how bugs modulate drugs: an educational initiative to explore the effects of human microbiome on drugs. BMC Bioinformatics 12 (Suppl. 7), A10 10.1186/1471-2105-12-S7-A10 - DOI
-
- Black P. N., DiRusso C. C., Metzger A. K., Heimert T. L. (1992). Cloning, sequencing, and expression of the fadD gene of Escherichia coli encoding acyl coenzyme A synthetase. J Biol Chem 267, 25513–25520. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

