Restriction of V3 region sequence divergence in the HIV-1 envelope gene during antiretroviral treatment in a cohort of recent seroconverters
- PMID: 23331949
- PMCID: PMC3605130
- DOI: 10.1186/1742-4690-10-8
Restriction of V3 region sequence divergence in the HIV-1 envelope gene during antiretroviral treatment in a cohort of recent seroconverters
Abstract
Background: Dynamic changes in Human Immunodeficiency Virus 1 (HIV-1) sequence diversity and divergence are associated with immune control during primary infection and progression to AIDS. Consensus sequencing or single genome amplification sequencing of the HIV-1 envelope (env) gene, in particular the variable (V) regions, is used as a marker for HIV-1 genome diversity, but population diversity is only minimally, or semi-quantitatively sampled using these methods.
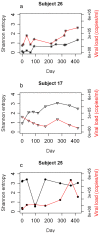
Results: Here we use second generation deep sequencing to determine inter-and intra-patient sequence heterogeneity and to quantify minor variants in a cohort of individuals either receiving or not receiving antiretroviral treatment following seroconversion; the SPARTAC trial. We show, through a cross-sectional study of sequence diversity of the env V3 in 30 antiretroviral-naive patients during primary infection that considerable population structure diversity exists, with some individuals exhibiting highly constrained plasma virus diversity. Diversity was independent of clinical markers (viral load, time from seroconversion, CD4 cell count) of infection. Serial sampling over 60 weeks of non-treated individuals that define three initially different diversity profiles showed that complex patterns of continuing HIV-1 sequence diversification and divergence could be readily detected. Evidence for minor sequence turnover, emergence of new variants and re-emergence of archived variants could be inferred from this analysis. Analysis of viral divergence over the same time period in patients who received short (12 weeks, ART12) or long course antiretroviral therapy (48 weeks, ART48) and a non-treated control group revealed that ART48 successfully suppressed viral divergence while ART12 did not have a significant effect.
Conclusions: Deep sequencing is a sensitive and reliable method for investigating the diversity of the env V3 as an important component of HIV-1 genome diversity. Detailed insights into the complex early intra-patient dynamics of env V3 diversity and divergence were explored in antiretroviral-naïve recent seroconverters. Long course antiretroviral therapy, initiated soon after seroconversion and administered for 48 weeks, restricts HIV-1 divergence significantly. The effect of ART12 and ART48 on clinical markers of HIV infection and progression is currently investigated in the SPARTAC trial.
Figures






References
-
- Keele BF, Giorgi EE, Salazar-Gonzalez JF, Decker JM, Pham KT, Salazar MG, Sun C, Grayson T, Wang S, Li H. et al.Identification and characterization of transmitted and early founder virus envelopes in primary HIV-1 infection. Proc Natl Acad Sci U S A. 2008;105:7552–7557. doi: 10.1073/pnas.0802203105. - DOI - PMC - PubMed
-
- Ganusov VV, Goonetilleke N, Liu MK, Ferrari G, Shaw GM, McMichael AJ, Borrow P, Korber BT, Perelson AS. Fitness costs and diversity of CTL response determine the rate of CTL escape during the acute and chronic phases of HIV infection. J Virol. 2011;85:10518–10528. doi: 10.1128/JVI.00655-11. - DOI - PMC - PubMed
-
- Love TM, Thurston SW, Keefer MC, Dewhurst S, Lee HY. Mathematical modeling of ultradeep sequencing data reveals that acute CD8+ T-lymphocyte responses exert strong selective pressure in simian immunodeficiency virus-infected macaques but still fail to clear founder epitope sequences. J Virol. 2010;84:5802–5814. doi: 10.1128/JVI.00117-10. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

