Conserved spatial organization of FG domains in the nuclear pore complex
- PMID: 23332057
- PMCID: PMC3540264
- DOI: 10.1016/j.bpj.2012.11.3823
Conserved spatial organization of FG domains in the nuclear pore complex
Abstract
Selective transport through the nuclear pore complex (NPC) requires nucleoporins containing natively unfolded phenylalanine-glycine (FG) domains. Several differing models for their dynamics within the pore have been proposed. We characterize the behavior of the FG nucleoporins in vivo using polarized fluorescence microscopy. Using nucleoporins tagged with green fluorescent protein along their FG domains, we show that some of these proteins are ordered, indicating an overall orientational organization within the NPC. This orientational ordering of the FG domains depends on their specific context within the NPC, but is independent of active transport and cargo load. For most nups, behavior does not depend on the FG motifs. These data support a model whereby local geometry constrains the orientational organization of the FG nups. Intriguingly, homologous yeast and mammalian proteins show conserved behavior, suggesting functional relevance. Our findings have implications for mechanistic models of NPC transport.
Copyright © 2013 Biophysical Society. Published by Elsevier Inc. All rights reserved.
Figures
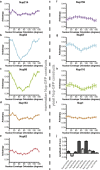

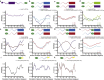

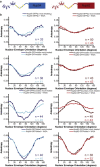

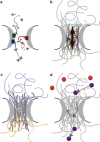
Comment in
-
Peptide ordering within nuclear pores in living cells.Biophys J. 2013 Jan 8;104(1):6-7. doi: 10.1016/j.bpj.2012.11.3824. Epub 2013 Jan 8. Biophys J. 2013. PMID: 23332053 Free PMC article. No abstract available.
References
-
- Ryan K.J., Wente S.R. The nuclear pore complex: a protein machine bridging the nucleus and cytoplasm. Curr. Opin. Cell Biol. 2000;12:361–371. - PubMed
-
- Moore M.S., Blobel G. The GTP-binding protein Ran/TC4 is required for protein import into the nucleus. Nature. 1993;365:661–663. - PubMed
-
- Bischoff F.R., Ponstingl H. Catalysis of guanine nucleotide exchange on Ran by the mitotic regulator RCC1. Nature. 1991;354:80–82. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

