Horizontal gene transfer and the evolution of bacterial and archaeal population structure
- PMID: 23332119
- PMCID: PMC3760709
- DOI: 10.1016/j.tig.2012.12.006
Horizontal gene transfer and the evolution of bacterial and archaeal population structure
Abstract
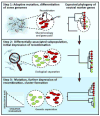
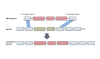
Many bacterial and archaeal lineages have a history of extensive and ongoing horizontal gene transfer and loss, as evidenced by the large differences in genome content even among otherwise closely related isolates. How ecologically cohesive populations might evolve and be maintained under such conditions of rapid gene turnover has remained controversial. Here we synthesize recent literature demonstrating the importance of habitat and niche in structuring horizontal gene transfer. This leads to a model of ecological speciation via gradual genetic isolation triggered by differential habitat-association of nascent populations. Further, we hypothesize that subpopulations can evolve through local gene-exchange networks by tapping into a gene pool that is adaptive towards local, continuously changing organismic interactions and is, to a large degree, responsible for the observed rapid gene turnover. Overall, these insights help to explain how bacteria and archaea form populations that display both ecological cohesion and high genomic diversity.
Copyright © 2012 Elsevier Ltd. All rights reserved.
Figures
References
-
- Tettelin H, et al. Comparative genomics: the bacterial pan-genome. Current Opinion in Microbiology. 2008;12:472–477. - PubMed
-
- Mira A, et al. The bacterial pan-genome:a new paradigm in microbiology. Int Microbiol. 2010;13:45–57. - PubMed
-
- Doolittle WF, Zhaxybayeva O. On the origin of prokaryotic species. Genome Research. 2009;19:744–756. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

