Spatial organization of ubiquitin ligase pathways orchestrates neuronal connectivity
- PMID: 23332798
- PMCID: PMC3622823
- DOI: 10.1016/j.tins.2012.12.004
Spatial organization of ubiquitin ligase pathways orchestrates neuronal connectivity
Abstract
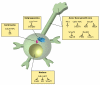
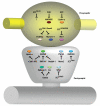
Recent studies have revealed that E3 ubiquitin ligases have essential functions in the establishment of neuronal circuits. Strikingly, a common emerging theme in these studies is that spatial organization of E3 ubiquitin ligases plays a critical role in the control of neuronal morphology and connectivity. E3 ubiquitin ligases localize to the nucleus, centrosome, Golgi apparatus, axon and dendrite cytoskeleton, and synapses in neurons. Localization of ubiquitin ligases within distinct subcellular compartments may facilitate neuronal responses to extrinsic cues and the ubiquitination of local substrates. Here, we review the functions of neuronal E3 ubiquitin ligases at distinct subcellular locales and explore how they regulate neuronal morphology and function in the nervous system.
Copyright © 2012 Elsevier Ltd. All rights reserved.
Figures

References
-
- Hicke L, Dunn R. Regulation of membrane protein transport by ubiquitin and ubiquitin-binding proteins. Annual review of cell and developmental biology. 2003;19:141–172. - PubMed
-
- Huang TT, D’Andrea AD. Regulation of DNA repair by ubiquitylation. Nat Rev Mol Cell Biol. 2006;7:323–334. - PubMed
-
- Kawabe H, Brose N. The role of ubiquitylation in nerve cell development. Nat Rev Neurosci. 2011;12:251–268. - PubMed
-
- Ang XL, Harper JW. Interwoven ubiquitination oscillators and control of cell cycle transitions. Sci STKE. 2004;2004:pe31. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

