PARP-1 mechanism for coupling DNA damage detection to poly(ADP-ribose) synthesis
- PMID: 23333033
- PMCID: PMC3572337
- DOI: 10.1016/j.sbi.2013.01.003
PARP-1 mechanism for coupling DNA damage detection to poly(ADP-ribose) synthesis
Abstract
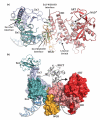
Poly(ADP-ribose) polymerase 1 (PARP-1) regulates gene transcription, cell death signaling, and DNA repair through production of the posttranslational modification poly(ADP-ribose). During the cellular response to genotoxic stress PARP-1 rapidly associates with DNA damage, which robustly stimulates poly(ADP-ribose) production over a low basal level of PARP-1 activity. DNA damage-dependent PARP-1 activity is central to understanding PARP-1 biological function, but structural insights into the mechanisms underlying this mode of regulation have remained elusive, in part due to the highly modular six-domain architecture of PARP-1. Recent structural studies have illustrated how PARP-1 uses specialized zinc fingers to detect DNA breaks through sequence-independent interaction with exposed nucleotide bases, a common feature of damaged and abnormal DNA structures. The mechanism of coupling DNA damage detection to elevated poly(ADP-ribose) production has been elucidated based on a crystal structure of the essential domains of PARP-1 in complex with a DNA strand break. The multiple domains of PARP-1 collapse onto damaged DNA, forming a network of interdomain contacts that introduce destabilizing alterations in the catalytic domain leading to an enhanced rate of poly(ADP-ribose) production.
Copyright © 2013. Published by Elsevier Ltd.
Figures


References
-
- Hassa PO, Hottiger MO. The diverse biological roles of mammalian PARPS, a small but powerful family of poly-ADP-ribose polymerases. Front Biosci. 2008;13:3046–3082. - PubMed
-
- Haince JF, McDonald D, Rodrigue A, Dery U, Masson JY, Hendzel MJ, Poirier GG. PARP1-dependent kinetics of recruitment of MRE11 and NBS1 proteins to multiple DNA damage sites. J Biol Chem. 2008;283:1197–1208. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

