A computational approach to developing mathematical models of polyploid meiosis
- PMID: 23335332
- PMCID: PMC3606088
- DOI: 10.1534/genetics.112.145581
A computational approach to developing mathematical models of polyploid meiosis
Abstract
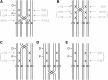
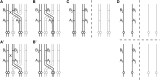
Mathematical models of meiosis that relate offspring to parental genotypes through parameters such as meiotic recombination frequency have been difficult to develop for polyploids. Existing models have limitations with respect to their analytic potential, their compatibility with insights into mechanistic aspects of meiosis, and their treatment of model parameters in terms of parameter dependencies. In this article I put forward a computational approach to the probabilistic modeling of meiosis. A computer program enumerates all possible paths through the phases of replication, pairing, recombination, and segregation, while keeping track of the probabilities of the paths according to the various parameters involved. Probabilities for classes of genotypes or phenotypes are added, and the resulting formulas are simplified by the symbolic-computation system Mathematica. An example application to autotetraploids results in a model that remedies the limitations of previous models mentioned above. In addition to the immediate implications, the computational approach presented here can be expected to be useful through opening avenues for modeling a host of processes, including meiosis in higher-order ploidies.
Figures

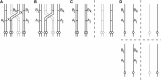
Similar articles
-
Analytical Methodology of Meiosis in Autopolyploid and Allopolyploid Plants.Methods Mol Biol. 2020;2061:141-168. doi: 10.1007/978-1-4939-9818-0_11. Methods Mol Biol. 2020. PMID: 31583658
-
Learning to tango with four (or more): the molecular basis of adaptation to polyploid meiosis.Plant Reprod. 2023 Mar;36(1):107-124. doi: 10.1007/s00497-022-00448-1. Epub 2022 Sep 23. Plant Reprod. 2023. PMID: 36149479 Free PMC article. Review.
-
Evolutionary consequences of polyploidy in prokaryotes and the origin of mitosis and meiosis.Biol Direct. 2016 Jun 8;11:28. doi: 10.1186/s13062-016-0131-8. Biol Direct. 2016. PMID: 27277956 Free PMC article.
-
A bivalent polyploid model for mapping quantitative trait loci in outcrossing tetraploids.Genetics. 2004 Jan;166(1):581-95. doi: 10.1534/genetics.166.1.581. Genetics. 2004. PMID: 15020446 Free PMC article.
-
Genetic regulation of meiosis in polyploid species: new insights into an old question.New Phytol. 2010 Apr;186(1):29-36. doi: 10.1111/j.1469-8137.2009.03084.x. Epub 2009 Nov 11. New Phytol. 2010. PMID: 19912546 Review.
Cited by
-
Linkage Analysis and Haplotype Phasing in Experimental Autopolyploid Populations with High Ploidy Level Using Hidden Markov Models.G3 (Bethesda). 2019 Oct 7;9(10):3297-3314. doi: 10.1534/g3.119.400378. G3 (Bethesda). 2019. PMID: 31405891 Free PMC article.
-
A two-locus model of selection in autotetraploids: Chromosomal gametic disequilibrium and selection for an adaptive epistatic gene combination.Heredity (Edinb). 2017 Nov;119(5):314-327. doi: 10.1038/hdy.2017.44. Epub 2017 Aug 23. Heredity (Edinb). 2017. PMID: 28832578 Free PMC article.
-
PERGOLA: fast and deterministic linkage mapping of polyploids.BMC Bioinformatics. 2017 Jan 4;18(1):12. doi: 10.1186/s12859-016-1416-8. BMC Bioinformatics. 2017. PMID: 28049428 Free PMC article.
-
Meiosis at three loci in autotetraploids: Probabilities of gamete modes and genotypes without and with preferential cross-over formation.Heredity (Edinb). 2023 Apr;130(4):223-235. doi: 10.1038/s41437-023-00595-9. Epub 2023 Feb 4. Heredity (Edinb). 2023. PMID: 36739333 Free PMC article.
References
-
- Brownfield L., Köhler C., 2011. Unreduced gamete formation in plants: mechanisms and prospects. J. Exp. Bot. 62(5): 1659–1668. - PubMed
-
- Comai L., 2005. The advantages and disadvantages of being polyploid. Nat. Rev. Genet. 6(11): 836–846. - PubMed
-
- Fisher R. A., 1947. The theory of linkage in polysomic inheritance. Philos. Trans. R. Soc. Lond. B Biol. Sci. 233(594): 55–87.
-
- Haynes K. G., Douches D. S., 1993. Estimation of the coefficient of double reduction in the cultivated tetraploid potato. Theor. Appl. Genet. 85(6): 857–862. - PubMed
-
- Jackson S. A., Iwata A., Lee S. H., Schmutz J., Shoemaker R., 2011. Sequencing crop genomes: approaches and applications. New Phytol. 191(4): 915–925. - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

