LATENT RANK CHANGE DETECTION FOR ANALYSIS OF SPLICE-JUNCTION MICROARRAYS WITH NONLINEAR EFFECTS
- PMID: 23335951
- PMCID: PMC3546815
- DOI: 10.1214/10-AOAS389SUPP
LATENT RANK CHANGE DETECTION FOR ANALYSIS OF SPLICE-JUNCTION MICROARRAYS WITH NONLINEAR EFFECTS
Abstract
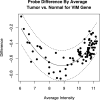
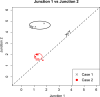
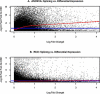
Alternative splicing of gene transcripts greatly expands the functional capacity of the genome, and certain splice isoforms may indicate specific disease states such as cancer. Splice junction microarrays interrogate thousands of splice junctions, but data analysis is difficult and error prone because of the increased complexity compared to differential gene expression analysis. We present Rank Change Detection (RCD) as a method to identify differential splicing events based upon a straightforward probabilistic model comparing the over- or underrepresentation of two or more competing isoforms. RCD has advantages over commonly used methods because it is robust to false positive errors due to nonlinear trends in microarray measurements. Further, RCD does not depend on prior knowledge of splice isoforms, yet it takes advantage of the inherent structure of mutually exclusive junctions, and it is conceptually generalizable to other types of splicing arrays or RNA-Seq. RCD specifically identifies the biologically important cases when a splice junction becomes more or less prevalent compared to other mutually exclusive junctions. The example data is from different cell lines of glioblastoma tumors assayed with Agilent microarrays.
Figures


References
-
- Cline M, Blume J, Cawley S, Clark T, Hu J, Lu G, Salomonis N, Wang H, Williams A. ANOSVA: A statistical method for detecting splice variation from expression data. Bioinformatics. 2005;21:I107–I115. - PubMed
-
- Cuperlovic-Culf M, Belacel N, Culf AS, Ouellette RJ. Microarray analysis of alternative splicing. Omics-A Journal of Integrative Biology. 2006;10:344–357. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
