High-resolution analysis and functional mapping of cleavage sites and substrate proteins of furin in the human proteome
- PMID: 23335997
- PMCID: PMC3545927
- DOI: 10.1371/journal.pone.0054290
High-resolution analysis and functional mapping of cleavage sites and substrate proteins of furin in the human proteome
Abstract
Background: There is a growing appreciation of the role of proteolytic processes in human health and disease, but tools for analysis of such processes on a proteome-wide scale are limited. Furin is a ubiquitous proprotein convertase that cleaves after basic residues and transforms secretory proproteins into biologically active proteins. Despite this important role, many furin substrates remain unknown in the human proteome.
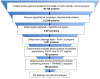
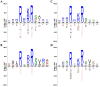
Methodology/principal findings: We devised an approach for proteinase target identification that combines an in silico discovery pipeline with highly multiplexed proteinase activity assays. We performed in silico analysis of the human proteome and identified over 1,050 secretory proteins as potential furin substrates. We then used a multiplexed protease assay to validate these tentative targets. The assay was carried out on over 3,260 overlapping peptides designed to represent P7-P1' and P4-P4' positions of furin cleavage sites in the candidate proteins. The obtained results greatly increased our knowledge of the unique cleavage preferences of furin, revealed the importance of both short-range (P4-P1) and long-range (P7-P6) interactions in defining furin cleavage specificity, demonstrated that the R-X-R/K/X-R ↓ motif alone is insufficient for predicting furin proteolysis of the substrate, and identified ≈ 490 potential protein substrates of furin in the human proteome.
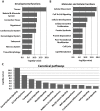
Conclusions/significance: The assignment of these substrates to cellular pathways suggests an important role of furin in development, including axonal guidance, cardiogenesis, and maintenance of stem cell pluripotency. The novel approach proposed in this study can be readily applied to other proteinases.
Conflict of interest statement
Figures

Similar articles
-
Substrate cleavage analysis of furin and related proprotein convertases. A comparative study.J Biol Chem. 2008 Jul 25;283(30):20897-906. doi: 10.1074/jbc.M803762200. Epub 2008 May 27. J Biol Chem. 2008. PMID: 18505722 Free PMC article.
-
X-ray Structures of the Proprotein Convertase Furin Bound with Substrate Analogue Inhibitors Reveal Substrate Specificity Determinants beyond the S4 Pocket.Biochemistry. 2018 Feb 13;57(6):925-934. doi: 10.1021/acs.biochem.7b01124. Epub 2018 Jan 26. Biochemistry. 2018. PMID: 29314830
-
FurinDB: A database of 20-residue furin cleavage site motifs, substrates and their associated drugs.Int J Mol Sci. 2011 Feb 8;12(2):1060-5. doi: 10.3390/ijms12021060. Int J Mol Sci. 2011. PMID: 21541042 Free PMC article.
-
Structure and function of eukaryotic proprotein processing enzymes of the subtilisin family of serine proteases.Crit Rev Oncog. 1993;4(2):115-36. Crit Rev Oncog. 1993. PMID: 8420571 Review.
-
Furin-mediated proprotein processing activity: involvement of negatively charged amino acid residues in the substrate binding region.Biochimie. 1994;76(3-4):210-6. doi: 10.1016/0300-9084(94)90148-1. Biochimie. 1994. PMID: 7819325 Review.
Cited by
-
SARS-CoV-2 S Mutations: A Lesson from the Viral World to Understand How Human Furin Works.Int J Mol Sci. 2023 Mar 1;24(5):4791. doi: 10.3390/ijms24054791. Int J Mol Sci. 2023. PMID: 36902222 Free PMC article.
-
SARS-CoV-2 and Emerging Variants: Unmasking Structure, Function, Infection, and Immune Escape Mechanisms.Front Cell Infect Microbiol. 2022 May 12;12:869832. doi: 10.3389/fcimb.2022.869832. eCollection 2022. Front Cell Infect Microbiol. 2022. PMID: 35646741 Free PMC article. Review.
-
Structures and dynamics of the novel S1/S2 protease cleavage site loop of the SARS-CoV-2 spike glycoprotein.J Struct Biol X. 2020;4:100038. doi: 10.1016/j.yjsbx.2020.100038. Epub 2020 Oct 5. J Struct Biol X. 2020. PMID: 33043289 Free PMC article.
-
Roles of host proteases in the entry of SARS-CoV-2.Anim Dis. 2023;3(1):12. doi: 10.1186/s44149-023-00075-x. Epub 2023 Apr 25. Anim Dis. 2023. PMID: 37128508 Free PMC article. Review.
-
Proprotein convertase FURIN constrains Th2 differentiation and is critical for host resistance against Toxoplasma gondii.J Immunol. 2014 Dec 1;193(11):5470-9. doi: 10.4049/jimmunol.1401629. Epub 2014 Oct 29. J Immunol. 2014. PMID: 25355923 Free PMC article.
References
-
- Seidah NG (2011) The proprotein convertases, 20 years later. Methods Mol Biol 768: 23–57. - PubMed
-
- Seidah NG (2011) What lies ahead for the proprotein convertases? Ann N Y Acad Sci 1220: 149–161. - PubMed
-
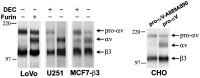
- Takahashi S, Nakagawa T, Kasai K, Banno T, Duguay SJ, et al. (1995) A second mutant allele of furin in the processing-incompetent cell line, LoVo. Evidence for involvement of the homo B domain in autocatalytic activation. J Biol Chem 270: 26565–26569. - PubMed
-
- Moehring JM, Inocencio NM, Robertson BJ, Moehring TJ (1993) Expression of mouse furin in a Chinese hamster cell resistant to Pseudomonas exotoxin A and viruses complements the genetic lesion. J Biol Chem 268: 2590–2594. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

