Trendspotting in the Protein Data Bank
- PMID: 23337870
- PMCID: PMC4068610
- DOI: 10.1016/j.febslet.2012.12.029
Trendspotting in the Protein Data Bank
Abstract
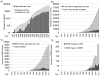
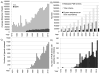
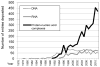
The Protein Data Bank (PDB) was established in 1971 as a repository for the three dimensional structures of biological macromolecules. Since then, more than 85000 biological macromolecule structures have been determined and made available in the PDB archive. Through analysis of the corpus of data, it is possible to identify trends that can be used to inform us abou the future of structural biology and to plan the best ways to improve the management of the ever-growing amount of PDB data.
Copyright © 2013 Federation of European Biochemical Societies. Published by Elsevier B.V. All rights reserved.
Figures








References
-
- Protein Data Bank. Protein Data Bank. Nature New Biol. 1971:233, 223.
-
- Berman H. The Protein Data Bank: a historical perspective. Acta Crystallogr A: Foundations of Crystallography. 2008;64:88–95. - PubMed
-
- International Union of Crystallography. Policy on publication and the deposition of data from crystallographic studies of biological macromolecules. Acta Cryst. 1989;A45:658.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

