Enhanced 5-methylcytosine detection in single-molecule, real-time sequencing via Tet1 oxidation
- PMID: 23339471
- PMCID: PMC3598637
- DOI: 10.1186/1741-7007-11-4
Enhanced 5-methylcytosine detection in single-molecule, real-time sequencing via Tet1 oxidation
Abstract
Background: DNA methylation serves as an important epigenetic mark in both eukaryotic and prokaryotic organisms. In eukaryotes, the most common epigenetic mark is 5-methylcytosine, whereas prokaryotes can have 6-methyladenine, 4-methylcytosine, or 5-methylcytosine. Single-molecule, real-time sequencing is capable of directly detecting all three types of modified bases. However, the kinetic signature of 5-methylcytosine is subtle, which presents a challenge for detection. We investigated whether conversion of 5-methylcytosine to 5-carboxylcytosine using the enzyme Tet1 would enhance the kinetic signature, thereby improving detection.
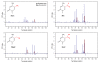
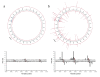
Results: We characterized the kinetic signatures of various cytosine modifications, demonstrating that 5-carboxylcytosine has a larger impact on the local polymerase rate than 5-methylcytosine. Using Tet1-mediated conversion, we show improved detection of 5-methylcytosine using in vitro methylated templates and apply the method to the characterization of 5-methylcytosine sites in the genomes of Escherichia coli MG1655 and Bacillus halodurans C-125.
Conclusions: We have developed a method for the enhancement of directly detecting 5-methylcytosine during single-molecule, real-time sequencing. Using Tet1 to convert 5-methylcytosine to 5-carboxylcytosine improves the detection rate of this important epigenetic marker, thereby complementing the set of readily detectable microbial base modifications, and enhancing the ability to interrogate eukaryotic epigenetic markers.
Figures


References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

