Custom oligonucleotide array-based CGH: a reliable diagnostic tool for detection of exonic copy-number changes in multiple targeted genes
- PMID: 23340513
- PMCID: PMC3746255
- DOI: 10.1038/ejhg.2012.279
Custom oligonucleotide array-based CGH: a reliable diagnostic tool for detection of exonic copy-number changes in multiple targeted genes
Abstract
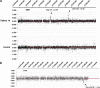
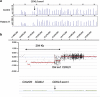
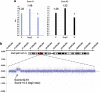
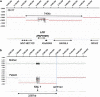
The frequency of disease-related large rearrangements (referred to as copy-number mutations, CNMs) varies among genes, and search for these mutations has an important place in diagnostic strategies. In recent years, CGH method using custom-designed high-density oligonucleotide-based arrays allowed the development of a powerful tool for detection of alterations at the level of exons and made it possible to provide flexibility through the possibility of modeling chips. The aim of our study was to test custom-designed oligonucleotide CGH array in a diagnostic laboratory setting that analyses several genes involved in various genetic diseases, and to compare it with conventional strategies. To this end, we designed a 12-plex CGH array (135k; 135 000 probes/subarray) (Roche Nimblegen) with exonic and intronic oligonucleotide probes covering 26 genes routinely analyzed in the laboratory. We tested control samples with known CNMs and patients for whom genetic causes underlying their disorders were unknown. The contribution of this technique is undeniable. Indeed, it appeared reproducible, reliable and sensitive enough to detect heterozygous single-exon deletions or duplications, complex rearrangements and somatic mosaicism. In addition, it improves reliability of CNM detection and allows determination of boundaries precisely enough to direct targeted sequencing of breakpoints. All of these points, associated with the possibility of a simultaneous analysis of several genes and scalability 'homemade' make it a valuable tool as a new diagnostic approach of CNMs.
Figures

Similar articles
-
Detection of exonic copy-number changes using a highly efficient oligonucleotide-based comparative genomic hybridization-array method.Hum Mutat. 2008 Sep;29(9):1083-90. doi: 10.1002/humu.20829. Hum Mutat. 2008. PMID: 18683213
-
Exon-level array CGH in a large clinical cohort demonstrates increased sensitivity of diagnostic testing for Mendelian disorders.Genet Med. 2012 Jun;14(6):594-603. doi: 10.1038/gim.2011.65. Epub 2012 Mar 1. Genet Med. 2012. PMID: 22382802
-
Detection of clinically relevant exonic copy-number changes by array CGH.Hum Mutat. 2010 Dec;31(12):1326-42. doi: 10.1002/humu.21360. Epub 2010 Nov 2. Hum Mutat. 2010. PMID: 20848651 Free PMC article.
-
Zoom-in array comparative genomic hybridization (aCGH) to detect germline rearrangements in cancer susceptibility genes.Methods Mol Biol. 2010;653:221-35. doi: 10.1007/978-1-60761-759-4_13. Methods Mol Biol. 2010. PMID: 20721746 Review.
-
Novel applications of array comparative genomic hybridization in molecular diagnostics.Expert Rev Mol Diagn. 2018 Jun;18(6):531-542. doi: 10.1080/14737159.2018.1479253. Epub 2018 May 31. Expert Rev Mol Diagn. 2018. PMID: 29848116 Review.
Cited by
-
NeuroArray, A Custom CGH Microarray to Decipher Copy Number Variants in Alzheimer's Disease.Curr Genomics. 2018 Sep;19(6):499-504. doi: 10.2174/1389202919666180122141425. Curr Genomics. 2018. PMID: 30258280 Free PMC article.
-
Microdeletion and microduplication analysis of chinese conotruncal defects patients with targeted array comparative genomic hybridization.PLoS One. 2013 Oct 2;8(10):e76314. doi: 10.1371/journal.pone.0076314. eCollection 2013. PLoS One. 2013. PMID: 24098474 Free PMC article.
-
EMQN best practice guidelines for genetic testing in dystrophinopathies.Eur J Hum Genet. 2020 Sep;28(9):1141-1159. doi: 10.1038/s41431-020-0643-7. Epub 2020 May 18. Eur J Hum Genet. 2020. PMID: 32424326 Free PMC article.
-
Improving molecular diagnosis of aniridia and WAGR syndrome using customized targeted array-based CGH.PLoS One. 2017 Feb 23;12(2):e0172363. doi: 10.1371/journal.pone.0172363. eCollection 2017. PLoS One. 2017. PMID: 28231309 Free PMC article.
-
Expert consensus document: European Consensus Statement on congenital hypogonadotropic hypogonadism--pathogenesis, diagnosis and treatment.Nat Rev Endocrinol. 2015 Sep;11(9):547-64. doi: 10.1038/nrendo.2015.112. Epub 2015 Jul 21. Nat Rev Endocrinol. 2015. PMID: 26194704 Review.
References
-
- Deburgrave N, Daoud F, Llense S, et al. Protein- and mRNA-based phenotype-genotype correlations in DMD/BMD with point mutations and molecular basis for BMD with nonsense and frameshift mutations in the DMD gene. Hum Mutat. 2007;28:183–195. - PubMed
-
- Tuffery-Giraud S, Beroud C, Leturcq F, et al. Genotype-phenotype analysis in 2,405 patients with a dystrophinopathy using the UMD-DMD database: a model of nationwide knowledgebase. Hum Mutat. 2009;30:934–945. - PubMed
-
- Ferec C, Casals T, Chuzhanova N, et al. Gross genomic rearrangements involving deletions in the CFTR gene: characterization of six new events from a large cohort of hitherto unidentified cystic fibrosis chromosomes and meta-analysis of the underlying mechanisms. Eur J Human Genet. 2006;14:567–576. - PubMed
-
- Casilli F, Di Rocco ZC, Gad S, et al. Rapid detection of novel BRCA1 rearrangements in high-risk breast-ovarian cancer families using multiplex PCR of short fluorescent fragments. Hum Mutat. 2002;20:218–226. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

