Autonomous bacterial localization and gene expression based on nearby cell receptor density
- PMID: 23340842
- PMCID: PMC3564257
- DOI: 10.1038/msb.2012.71
Autonomous bacterial localization and gene expression based on nearby cell receptor density
Abstract
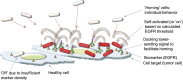
Escherichia coli were genetically modified to enable programmed motility, sensing, and actuation based on the density of features on nearby surfaces. Then, based on calculated feature density, these cells expressed marker proteins to indicate phenotypic response. Specifically, site-specific synthesis of bacterial quorum sensing autoinducer-2 (AI-2) is used to initiate and recruit motile cells. In our model system, we rewired E. coli's AI-2 signaling pathway to direct bacteria to a squamous cancer cell line of head and neck (SCCHN), where they initiate synthesis of a reporter (drug surrogate) based on a threshold density of epidermal growth factor receptor (EGFR). This represents a new type of controller for targeted drug delivery as actuation (synthesis and delivery) depends on a receptor density marking the diseased cell. The ability to survey local surfaces and initiate gene expression based on feature density represents a new area-based switch in synthetic biology that will find use beyond the proposed cancer model here.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures




References
-
- Anderson JC, Clarke EJ, Arkin AP, Voigt CA (2006) Environmentally controlled invasion of cancer cells by engineered bacteria. J Mol Biol 355: 619–627 - PubMed
-
- Atsumi S, Hanai T, Liao JC (2008) Non-fermentative pathways for synthesis of branched-chain higher alcohols as biofuels. Nature 451: 86–89 - PubMed
-
- Bentley WE, Mirjalili N, Andersen DC, Davis RH, Kompala DS (1990) Plasmid-encoded protein: the principal factor in the "metabolic burden" associated with recombinant bacteria. Biotechnol Bioeng 35: 668–681 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

