Genome-scale engineering for systems and synthetic biology
- PMID: 23340847
- PMCID: PMC3564264
- DOI: 10.1038/msb.2012.66
Genome-scale engineering for systems and synthetic biology
Abstract
Genome-modification technologies enable the rational engineering and perturbation of biological systems. Historically, these methods have been limited to gene insertions or mutations at random or at a few pre-defined locations across the genome. The handful of methods capable of targeted gene editing suffered from low efficiencies, significant labor costs, or both. Recent advances have dramatically expanded our ability to engineer cells in a directed and combinatorial manner. Here, we review current technologies and methodologies for genome-scale engineering, discuss the prospects for extending efficient genome modification to new hosts, and explore the implications of continued advances toward the development of flexibly programmable chasses, novel biochemistries, and safer organismal and ecological engineering.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures
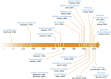


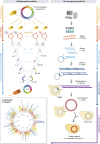
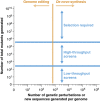

References
-
- Agarwal KL, Yamazaki A, Cashion PJ, Khorana HG (1972) Chemical synthesis of polynucleotides. Angew Chem Int Ed Engl 11: 451–459 - PubMed
-
- Albert H, Dale EC, Lee E, Ow DW (1995) Site-specific integration of DNA into wild-type and mutant lox sites placed in the plant genome. Plant J 7: 649–659 - PubMed
-
- Alper H, Miyaoku K, Stephanopoulos G (2005) Construction of lycopene-overproducing E. coli strains by combining systematic and combinatorial gene knockout targets. Nat Biotechnol 23: 612–616 - PubMed
-
- Alper H, Stephanopoulos G (2007) Global transcription machinery engineering: a new approach for improving cellular phenotype. Metab Eng 9: 258–267 - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

