Turning gold into 'junk': transposable elements utilize central proteins of cellular networks
- PMID: 23341038
- PMCID: PMC3597677
- DOI: 10.1093/nar/gkt011
Turning gold into 'junk': transposable elements utilize central proteins of cellular networks
Abstract
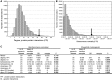
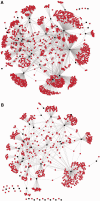
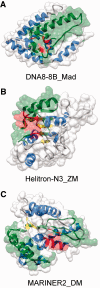
The numerous discovered cases of domesticated transposable element (TE) proteins led to the recognition that TEs are a significant source of evolutionary innovation. However, much less is known about the reverse process, whether and to what degree the evolution of TEs is influenced by the genome of their hosts. We addressed this issue by searching for cases of incorporation of host genes into the sequence of TEs and examined the systems-level properties of these genes using the Saccharomyces cerevisiae and Drosophila melanogaster genomes. We identified 51 cases where the evolutionary scenario was the incorporation of a host gene fragment into a TE consensus sequence, and we show that both the yeast and fly homologues of the incorporated protein sequences have central positions in the cellular networks. An analysis of selective pressure (Ka/Ks ratio) detected significant selection in 37% of the cases. Recent research on retrovirus-host interactions shows that virus proteins preferentially target hubs of the host interaction networks enabling them to take over the host cell using only a few proteins. We propose that TEs face a similar evolutionary pressure to evolve proteins with high interacting capacities and take some of the necessary protein domains directly from their hosts.
Figures


References
-
- Volff J. Turning junk into gold: domestication of transposable elements and the creation of new genes in eukaryotes. Bioessays. 2006;28:913–922. - PubMed
-
- Jurka J, Kapitonov VV, Kohany O, Jurka MV. Repetitive sequences in complex genomes: structure and evolution. Annu. Rev. Genomics. Hum. Genet. 2007;8:241–259. 10.1146/annurev.genom.8.080706.092416. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

