Orchestration of thiamin biosynthesis and central metabolism by combined action of the thiamin pyrophosphate riboswitch and the circadian clock in Arabidopsis
- PMID: 23341335
- PMCID: PMC3584542
- DOI: 10.1105/tpc.112.106385
Orchestration of thiamin biosynthesis and central metabolism by combined action of the thiamin pyrophosphate riboswitch and the circadian clock in Arabidopsis
Abstract
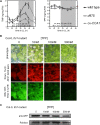
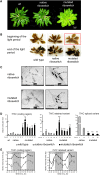
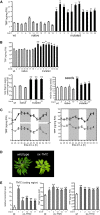
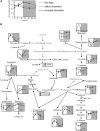
Riboswitches are natural RNA elements that posttranscriptionally regulate gene expression by binding small molecules and thereby autonomously control intracellular levels of these metabolites. Although riboswitch-based mechanisms have been examined extensively, the integration of their activity with global physiology and metabolism has been largely overlooked. Here, we explored the regulation of thiamin biosynthesis and the consequences of thiamin pyrophosphate riboswitch deficiency on metabolism in Arabidopsis thaliana. Our results show that thiamin biosynthesis is largely regulated by the circadian clock via the activity of the THIAMIN C SYNTHASE (THIC) promoter, while the riboswitch located at the 3' untranslated region of this gene controls overall thiamin biosynthesis. Surprisingly, the results also indicate that the rate of thiamin biosynthesis directs the activity of thiamin-requiring enzymes and consecutively determines the rate of carbohydrate oxidation via the tricarboxylic acid cycle and pentose-phosphate pathway. Our model suggests that in Arabidopsis, the THIC promoter and the thiamin-pyrophosphate riboswitch act simultaneously to tightly regulate thiamin biosynthesis in a circadian manner and consequently sense and control vital points of core cellular metabolism.
Figures



Similar articles
-
Appropriate Thiamin Pyrophosphate Levels Are Required for Acclimation to Changes in Photoperiod.Plant Physiol. 2019 May;180(1):185-197. doi: 10.1104/pp.18.01346. Epub 2019 Mar 5. Plant Physiol. 2019. PMID: 30837347 Free PMC article.
-
Riboswitch-dependent gene regulation and its evolution in the plant kingdom.Genes Dev. 2007 Nov 15;21(22):2874-9. doi: 10.1101/gad.443907. Genes Dev. 2007. PMID: 18006684 Free PMC article.
-
Analysis of Chlamydomonas thiamin metabolism in vivo reveals riboswitch plasticity.Proc Natl Acad Sci U S A. 2013 Sep 3;110(36):14622-7. doi: 10.1073/pnas.1307741110. Epub 2013 Aug 19. Proc Natl Acad Sci U S A. 2013. PMID: 23959877 Free PMC article.
-
Small molecules that interact with RNA: riboswitch-based gene control and its involvement in metabolic regulation in plants and algae.Plant J. 2014 Aug;79(4):693-703. doi: 10.1111/tpj.12540. Epub 2014 Jun 17. Plant J. 2014. PMID: 24773387 Review.
-
Switching the light on plant riboswitches.Trends Plant Sci. 2008 Oct;13(10):526-33. doi: 10.1016/j.tplants.2008.07.004. Epub 2008 Sep 6. Trends Plant Sci. 2008. PMID: 18778966 Review.
Cited by
-
On the nature of thiamine triphosphate in Arabidopsis.Plant Direct. 2020 Aug 30;4(8):e00258. doi: 10.1002/pld3.258. eCollection 2020 Aug. Plant Direct. 2020. PMID: 32885135 Free PMC article.
-
B vitamin supply in plants and humans: the importance of vitamer homeostasis.Plant J. 2022 Aug;111(3):662-682. doi: 10.1111/tpj.15859. Epub 2022 Jun 27. Plant J. 2022. PMID: 35673947 Free PMC article. Review.
-
Salvage of the thiamin pyrimidine moiety by plant TenA proteins lacking an active-site cysteine.Biochem J. 2014 Oct 1;463(1):145-55. doi: 10.1042/BJ20140522. Biochem J. 2014. PMID: 25014715 Free PMC article.
-
Metabolic engineering of rice endosperm towards higher vitamin B1 accumulation.Plant Biotechnol J. 2021 Jun;19(6):1253-1267. doi: 10.1111/pbi.13545. Epub 2021 Feb 11. Plant Biotechnol J. 2021. PMID: 33448624 Free PMC article.
-
Examining strategies to facilitate vitamin B1 biofortification of plants by genetic engineering.Front Plant Sci. 2013 May 29;4:160. doi: 10.3389/fpls.2013.00160. eCollection 2013. Front Plant Sci. 2013. PMID: 23755056 Free PMC article.
References
-
- Ajjawi I., Rodriguez Milla M.A., Cushman J., Shintani D.K. (2007b). Thiamin pyrophosphokinase is required for thiamin cofactor activation in Arabidopsis. Plant Mol. Biol. 65: 151–162 - PubMed
-
- Ajjawi I., Tsegaye Y., Shintani D. (2007a). Determination of the genetic, molecular, and biochemical basis of the Arabidopsis thaliana thiamin auxotroph th1. Arch. Biochem. Biophys. 459: 107–114 - PubMed
-
- Alabadí D., Oyama T., Yanovsky M.J., Harmon F.G., Más P., Kay S.A. (2001). Reciprocal regulation between TOC1 and LHY/CCA1 within the Arabidopsis circadian clock. Science 293: 880–883 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

