Organ evolution in angiosperms driven by correlated divergences of gene sequences and expression patterns
- PMID: 23341336
- PMCID: PMC3584551
- DOI: 10.1105/tpc.112.106716
Organ evolution in angiosperms driven by correlated divergences of gene sequences and expression patterns
Abstract
The evolution of a species involves changes in its genome and its transcriptome. Divergence in expression patterns may be more important than divergence in sequences for determining phenotypic changes, particularly among closely related species. We examined the relationships between organ evolution, sequence evolution, and expression evolution in Arabidopsis thaliana, rice (Oryza sativa), and maize (Zea mays). We found correlated divergence of gene sequences and expression patterns, with distinct divergence rates that depend on the organ types in which a gene is expressed. For instance, genes specifically expressed in reproductive organs (i.e., stamen) evolve more quickly than those specifically expressed in vegetative organs (e.g., root). The different rates in organ evolution may be due to different degrees of functional constraint associated with the different physiological functions of plant organs. Additionally, the evolutionary rate of a gene sequence is correlated with the breadth of its expression in terms of the number of tissues, the number of coregulation modules, and the number of species in which the gene is expressed, as well as the number of genes with which it may interact. This linkage supports the hypothesis that constitutively expressed genes may experience higher levels of functional constraint accumulated from multiple tissues than do tissue-specific genes.
Figures
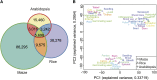

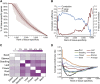

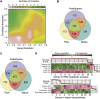
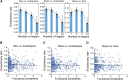
Comment in
-
Gene expression in angiosperm organ evolution.Plant Cell. 2013 Feb;25(2):357. doi: 10.1105/tpc.113.250210. Epub 2013 Feb 12. Plant Cell. 2013. PMID: 23404885 Free PMC article. No abstract available.
Similar articles
-
Contrasting transcriptional responses of PYR1/PYL/RCAR ABA receptors to ABA or dehydration stress between maize seedling leaves and roots.BMC Plant Biol. 2016 Apr 21;16:99. doi: 10.1186/s12870-016-0764-x. BMC Plant Biol. 2016. PMID: 27101806 Free PMC article.
-
ZINC-INDUCED FACILITATOR-LIKE family in plants: lineage-specific expansion in monocotyledons and conserved genomic and expression features among rice (Oryza sativa) paralogs.BMC Plant Biol. 2011 Jan 25;11:20. doi: 10.1186/1471-2229-11-20. BMC Plant Biol. 2011. PMID: 21266036 Free PMC article.
-
Evolution and divergence of SBP-box genes in land plants.BMC Genomics. 2015 Oct 14;16:787. doi: 10.1186/s12864-015-1998-y. BMC Genomics. 2015. PMID: 26467431 Free PMC article.
-
Combining expression and comparative evolutionary analysis. The COBRA gene family.Plant Physiol. 2007 Jan;143(1):172-87. doi: 10.1104/pp.106.087262. Epub 2006 Nov 10. Plant Physiol. 2007. PMID: 17098858 Free PMC article.
-
Plant organelle proteomics: collaborating for optimal cell function.Mass Spectrom Rev. 2011 Sep-Oct;30(5):772-853. doi: 10.1002/mas.20301. Epub 2010 Oct 29. Mass Spectrom Rev. 2011. PMID: 21038434 Review.
Cited by
-
Expression divergence of expansin genes drive the heteroblasty in Ceratopteris chingii.BMC Biol. 2023 Nov 6;21(1):244. doi: 10.1186/s12915-023-01743-7. BMC Biol. 2023. PMID: 37926805 Free PMC article.
-
Pan-organ transcriptome variation across 21 cancer types.Oncotarget. 2017 Jan 24;8(4):6809-6818. doi: 10.18632/oncotarget.14303. Oncotarget. 2017. PMID: 28036280 Free PMC article.
-
UPLC/Q-TOF MS-based metabolomics and qRT-PCR in enzyme gene screening with key role in triterpenoid saponin biosynthesis of Polygala tenuifolia.PLoS One. 2014 Aug 22;9(8):e105765. doi: 10.1371/journal.pone.0105765. eCollection 2014. PLoS One. 2014. PMID: 25148032 Free PMC article.
-
Gene expression in angiosperm organ evolution.Plant Cell. 2013 Feb;25(2):357. doi: 10.1105/tpc.113.250210. Epub 2013 Feb 12. Plant Cell. 2013. PMID: 23404885 Free PMC article. No abstract available.
-
Plant organ evolution revealed by phylotranscriptomics in Arabidopsis thaliana.Sci Rep. 2017 Aug 8;7(1):7567. doi: 10.1038/s41598-017-07866-6. Sci Rep. 2017. PMID: 28790409 Free PMC article.
References
-
- Bergmann, S., Ihmels, J., and Barkai, N. (2003). Iterative signature algorithm for the analysis of large-scale gene expression data. Phys. Rev. E Stat Nonlin. Soft Matter Phys. 67: 031902. - PubMed
-
- Brawand D., et al. (2011). The evolution of gene expression levels in mammalian organs. Nature 478: 343–348 - PubMed
-
- Cao J., et al. (2011). Whole-genome sequencing of multiple Arabidopsis thaliana populations. Nat. Genet. 43: 956–963 - PubMed
-
- Clark N.L., Aagaard J.E., Swanson W.J. (2006). Evolution of reproductive proteins from animals and plants. Reproduction 131: 11–22 - PubMed
-
- Csárdi G., Kutalik Z., Bergmann S. (2010). Modular analysis of gene expression data with R. Bioinformatics 26: 1376–1377 - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

