The polysialyltransferases interact with sequences in two domains of the neural cell adhesion molecule to allow its polysialylation
- PMID: 23341449
- PMCID: PMC3591636
- DOI: 10.1074/jbc.M112.438374
The polysialyltransferases interact with sequences in two domains of the neural cell adhesion molecule to allow its polysialylation
Abstract
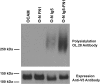
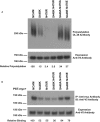
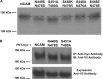
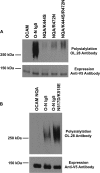
The neural cell adhesion molecule (NCAM) is the major substrate for the polysialyltransferases (polySTs), ST8SiaII/STX and ST8SiaIV/PST. The polysialylation of NCAM N-glycans decreases cell adhesion and alters signaling. Previous work demonstrated that the first fibronectin type III repeat (FN1) of NCAM is required for polyST recognition and the polysialylation of the N-glycans on the adjacent Ig5 domain. In this work, we highlight the importance of an FN1 acidic patch in polyST recognition and also reveal that the polySTs are required to interact with sequences in the Ig5 domain for polysialylation to occur. We find that features of the Ig5 domain of the olfactory cell adhesion molecule (OCAM) are responsible for its lack of polysialylation. Specifically, two basic OCAM Ig5 residues (Lys and Arg) found near asparagines equivalent to those carrying the polysialylated N-glycans in NCAM substantially decrease or eliminate polysialylation when used to replace the smaller and more neutral residues (Ser and Asn) in analogous positions in NCAM Ig5. This decrease in polysialylation does not reflect altered glycosylation but instead is correlated with a decrease in polyST-NCAM binding. In addition, inserting non-conserved OCAM sequences into NCAM Ig5, including an "extra" N-glycosylation site, decreases or completely blocks NCAM polysialylation. Taken together, these results indicate that the polySTs not only recognize an acidic patch in the FN1 domain of NCAM but also must contact sequences in the Ig5 domain for polysialylation of Ig5 N-glycans to occur.
Figures


Similar articles
-
Sequences from the first fibronectin type III repeat of the neural cell adhesion molecule allow O-glycan polysialylation of an adhesion molecule chimera.J Biol Chem. 2010 Nov 5;285(45):35056-67. doi: 10.1074/jbc.M110.170209. Epub 2010 Aug 30. J Biol Chem. 2010. PMID: 20805222 Free PMC article.
-
Sequences at the interface of the fifth immunoglobulin domain and first fibronectin type III repeat of the neural cell adhesion molecule are critical for its polysialylation.J Biol Chem. 2011 Feb 11;286(6):4525-34. doi: 10.1074/jbc.M110.200386. Epub 2010 Dec 3. J Biol Chem. 2011. PMID: 21131353 Free PMC article.
-
Specific amino acids in the first fibronectin type III repeat of the neural cell adhesion molecule play a role in its recognition and polysialylation by the polysialyltransferase ST8Sia IV/PST.J Biol Chem. 2005 Sep 16;280(37):32340-8. doi: 10.1074/jbc.M506217200. Epub 2005 Jul 18. J Biol Chem. 2005. PMID: 16027151
-
3D structural conformation and functional domains of polysialyltransferase ST8Sia IV required for polysialylation of neural cell adhesion molecules.Protein Pept Lett. 2015;22(2):137-48. doi: 10.2174/0929866521666141019192221. Protein Pept Lett. 2015. PMID: 25329332 Review.
-
A Possible Modulation Mechanism of Intramolecular and Intermolecular Interactions for NCAM Polysialylation and Cell Migration.Curr Top Med Chem. 2019;19(25):2271-2282. doi: 10.2174/1568026619666191018094805. Curr Top Med Chem. 2019. PMID: 31648641 Review.
Cited by
-
Glycobiology of neuroblastoma: impact on tumor behavior, prognosis, and therapeutic strategies.Front Oncol. 2014 May 23;4:114. doi: 10.3389/fonc.2014.00114. eCollection 2014. Front Oncol. 2014. PMID: 24904828 Free PMC article. Review.
-
Sialylation of N-glycans: mechanism, cellular compartmentalization and function.Histochem Cell Biol. 2017 Feb;147(2):149-174. doi: 10.1007/s00418-016-1520-x. Epub 2016 Dec 14. Histochem Cell Biol. 2017. PMID: 27975143 Free PMC article. Review.
-
Autopolysialylation of polysialyltransferases is required for polysialylation and polysialic acid chain elongation on select glycoprotein substrates.J Biol Chem. 2018 Jan 12;293(2):701-716. doi: 10.1074/jbc.RA117.000401. Epub 2017 Nov 28. J Biol Chem. 2018. PMID: 29183999 Free PMC article.
-
Structure of human ST8SiaIII sialyltransferase provides insight into cell-surface polysialylation.Nat Struct Mol Biol. 2015 Aug;22(8):627-35. doi: 10.1038/nsmb.3060. Epub 2015 Jul 20. Nat Struct Mol Biol. 2015. PMID: 26192331
-
NCAM and attached polysialic acid affect behaviors of breast epithelial cells through differential signaling pathways.Acta Biochim Biophys Sin (Shanghai). 2024 Oct 15;56(11):1584-1593. doi: 10.3724/abbs.2024176. Acta Biochim Biophys Sin (Shanghai). 2024. PMID: 39420834 Free PMC article.
References
-
- Haltiwanger R. S., Lowe J. B. (2004) Role of glycosylation in development. Annu. Rev. Biochem. 73, 491–537 - PubMed
-
- Johnson C. P., Fujimoto I., Rutishauser U., Leckband D. E. (2005) Direct evidence that neural cell adhesion molecule (NCAM) polysialylation increases intermembrane repulsion and abrogates adhesion. J. Biol. Chem. 280, 137–145 - PubMed
-
- Hildebrandt H., Mühlenhoff M., Weinhold B., Gerardy-Schahn R. (2007) Dissecting polysialic acid and NCAM functions in brain development. J. Neurochem. 103, 56–64 - PubMed
-
- Rutishauser U. (2008) Polysialic acid in the plasticity of the developing and adult vertebrate nervous system. Nat. Rev. Neurosci. 9, 26–35 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

