The molecular mechanism of Hsp100 chaperone inhibition by the prion curing agent guanidinium chloride
- PMID: 23341453
- PMCID: PMC3591616
- DOI: 10.1074/jbc.M112.432583
The molecular mechanism of Hsp100 chaperone inhibition by the prion curing agent guanidinium chloride
Abstract
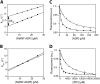
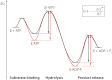
The Hsp100 chaperones ClpB and Hsp104 utilize the energy from ATP hydrolysis to reactivate aggregated proteins in concert with the DnaK/Hsp70 chaperone system, thereby playing an important role in protein quality control. They belong to the family of AAA+ proteins (ATPases associated with various cellular activities), possess two nucleotide binding domains per monomer (NBD1 and NBD2), and oligomerize into hexameric ring complexes. Furthermore, Hsp104 is involved in yeast prion propagation and inheritance. It is well established that low concentrations of guanidinium chloride (GdmCl) inhibit the ATPase activity of Hsp104, leading to so called "prion curing," the loss of prion-related phenotypes. Here, we present mechanistic details about the Hsp100 chaperone inhibition by GdmCl using the Hsp104 homolog ClpB from Thermus thermophilus. Initially, we demonstrate that NBD1 of ClpB, which was previously considered inactive as a separately expressed construct, is a fully active ATPase on its own. Next, we show that only NBD1, but not NBD2, is affected by GdmCl. We present a crystal structure of ClpB NBD1 in complex with GdmCl and ADP, showing that the Gdm(+) ion binds specifically to the active site of NBD1. A conserved essential glutamate residue is involved in this interaction. Additionally, Gdm(+) interacts directly with the nucleotide, thereby increasing the nucleotide binding affinity of NBD1. We propose that both the interference with the essential glutamate and the modulation of nucleotide binding properties in NBD1 is responsible for the GdmCl-specific inhibition of Hsp100 chaperones.
Figures





Similar articles
-
trans-Acting arginine residues in the AAA+ chaperone ClpB allosterically regulate the activity through inter- and intradomain communication.J Biol Chem. 2014 Nov 21;289(47):32965-76. doi: 10.1074/jbc.M114.608828. Epub 2014 Sep 24. J Biol Chem. 2014. PMID: 25253689 Free PMC article.
-
Analysis of the cooperative ATPase cycle of the AAA+ chaperone ClpB from Thermus thermophilus by using ordered heterohexamers with an alternating subunit arrangement.J Biol Chem. 2015 Apr 10;290(15):9789-800. doi: 10.1074/jbc.M114.617696. Epub 2015 Feb 24. J Biol Chem. 2015. PMID: 25713084 Free PMC article.
-
Crystal structure of E. coli Hsp100 ClpB nucleotide-binding domain 1 (NBD1) and mechanistic studies on ClpB ATPase activity.J Mol Biol. 2002 May 10;318(4):1127-37. doi: 10.1016/S0022-2836(02)00188-2. J Mol Biol. 2002. PMID: 12054807
-
The ClpB/Hsp104 molecular chaperone-a protein disaggregating machine.J Struct Biol. 2004 Apr-May;146(1-2):99-105. doi: 10.1016/j.jsb.2003.11.016. J Struct Biol. 2004. PMID: 15037241 Review.
-
Towards a unifying mechanism for ClpB/Hsp104-mediated protein disaggregation and prion propagation.Biochem Cell Biol. 2010 Feb;88(1):63-75. doi: 10.1139/o09-118. Biochem Cell Biol. 2010. PMID: 20130680 Review.
Cited by
-
SMR peptide antagonizes Staphylococcus aureus biofilm formation.Microbiol Spectr. 2024 Feb 6;12(2):e0258323. doi: 10.1128/spectrum.02583-23. Epub 2024 Jan 3. Microbiol Spectr. 2024. PMID: 38170991 Free PMC article.
-
Structural basis for the disaggregase activity and regulation of Hsp104.Elife. 2016 Nov 30;5:e21516. doi: 10.7554/eLife.21516. Elife. 2016. PMID: 27901467 Free PMC article.
-
Cooperation of Hsp70 and Hsp100 chaperone machines in protein disaggregation.Front Mol Biosci. 2015 May 19;2:22. doi: 10.3389/fmolb.2015.00022. eCollection 2015. Front Mol Biosci. 2015. PMID: 26042222 Free PMC article. Review.
-
Guanidinium export is the primal function of SMR family transporters.Proc Natl Acad Sci U S A. 2018 Mar 20;115(12):3060-3065. doi: 10.1073/pnas.1719187115. Epub 2018 Mar 5. Proc Natl Acad Sci U S A. 2018. PMID: 29507227 Free PMC article.
-
Guanidine hydrochloride reactivates an ancient septin hetero-oligomer assembly pathway in budding yeast.Elife. 2020 Jan 28;9:e54355. doi: 10.7554/eLife.54355. Elife. 2020. PMID: 31990274 Free PMC article.
References
-
- Sanchez Y., Lindquist S. L. (1990) HSP104 required for induced thermotolerance. Science 248, 1112–1115 - PubMed
-
- Glover J. R., Lindquist S. (1998) Hsp104, Hsp70, and Hsp40: a novel chaperone system that rescues previously aggregated proteins. Cell 94, 73–82 - PubMed
-
- Tucker P. A., Sallai L. (2007) The AAA+ superfamily—a myriad of motions. Curr. Opin. Struct. Biol. 17, 641–652 - PubMed
-
- Hanson P. I., Whiteheart S. W. (2005) AAA+ proteins: have engine, will work. Nat. Rev. Mol. Cell Biol. 6, 519–529 - PubMed
-
- Weibezahn J., Tessarz P., Schlieker C., Zahn R., Maglica Z., Lee S., Zentgraf H., Weber-Ban E. U., Dougan D. A., Tsai F. T., Mogk A., Bukau B. (2004) Thermotolerance requires refolding of aggregated proteins by substrate translocation through the central pore of ClpB. Cell 119, 653–665 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

