The HsiB1C1 (TssB-TssC) complex of the Pseudomonas aeruginosa type VI secretion system forms a bacteriophage tail sheathlike structure
- PMID: 23341461
- PMCID: PMC3597794
- DOI: 10.1074/jbc.M112.439273
The HsiB1C1 (TssB-TssC) complex of the Pseudomonas aeruginosa type VI secretion system forms a bacteriophage tail sheathlike structure
Abstract
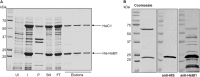
Protein secretion systems in Gram-negative bacteria evolved into a variety of molecular nanomachines. They are related to cell envelope complexes, which are involved in assembly of surface appendages or transport of solutes. They are classified as types, the most recent addition being the type VI secretion system (T6SS). The T6SS displays similarities to bacteriophage tail, which drives DNA injection into bacteria. The Hcp protein is related to the T4 bacteriophage tail tube protein gp19, whereas VgrG proteins structurally resemble the gp27/gp5 puncturing device of the phage. The tube and spike of the phage are pushed through the bacterial envelope upon contraction of a tail sheath composed of gp18. In Vibrio cholerae it was proposed that VipA and VipB assemble into a tail sheathlike structure. Here we confirm these previous data by showing that HsiB1 and HsiC1 of the Pseudomonas aeruginosa H1-T6SS assemble into tubules resulting from stacking of cogwheel-like structures showing predominantly 12-fold symmetry. The internal diameter of the cogwheels is ~100 Å, which is large enough to accommodate an Hcp tube whose external diameter has been reported to be 85 Å. The N-terminal 212 residues of HsiC1 are sufficient to form a stable complex with HsiB1, but the C terminus of HsiC1 is essential for the formation of the tubelike structure. Bioinformatics analysis suggests that HsiC1 displays similarities to gp18-like proteins in its C-terminal region. In conclusion, we provide further structural and mechanistic insights into the T6SS and show that a phage sheathlike structure is likely to be a conserved element across all T6SSs.
Figures






Similar articles
-
Coevolution of the ATPase ClpV, the sheath proteins TssB and TssC, and the accessory protein TagJ/HsiE1 distinguishes type VI secretion classes.J Biol Chem. 2014 Nov 21;289(47):33032-43. doi: 10.1074/jbc.M114.600510. Epub 2014 Oct 10. J Biol Chem. 2014. PMID: 25305017 Free PMC article.
-
The archetype Pseudomonas aeruginosa proteins TssB and TagJ form a novel subcomplex in the bacterial type VI secretion system.Mol Microbiol. 2012 Oct;86(2):437-56. doi: 10.1111/j.1365-2958.2012.08204.x. Epub 2012 Aug 29. Mol Microbiol. 2012. PMID: 22906320
-
Atomic Structure of Type VI Contractile Sheath from Pseudomonas aeruginosa.Structure. 2018 Feb 6;26(2):329-336.e3. doi: 10.1016/j.str.2017.12.005. Epub 2018 Jan 4. Structure. 2018. PMID: 29307484 Free PMC article.
-
Tubules and donuts: a type VI secretion story.Mol Microbiol. 2010 May;76(4):815-21. doi: 10.1111/j.1365-2958.2010.07171.x. Epub 2010 Apr 23. Mol Microbiol. 2010. PMID: 20444095 Review.
-
Composition, function, and regulation of T6SS in Pseudomonas aeruginosa.Microbiol Res. 2015 Mar;172:19-25. doi: 10.1016/j.micres.2015.01.004. Epub 2015 Jan 7. Microbiol Res. 2015. PMID: 25721475 Review.
Cited by
-
Recruitment of Mobile Genetic Elements for Diverse Cellular Functions in Prokaryotes.Front Mol Biosci. 2022 Mar 24;9:821197. doi: 10.3389/fmolb.2022.821197. eCollection 2022. Front Mol Biosci. 2022. PMID: 35402511 Free PMC article. Review.
-
Dissection of the TssB-TssC interface during type VI secretion sheath complex formation.PLoS One. 2013 Nov 25;8(11):e81074. doi: 10.1371/journal.pone.0081074. eCollection 2013. PLoS One. 2013. PMID: 24282569 Free PMC article.
-
Genetically distinct pathways guide effector export through the type VI secretion system.Mol Microbiol. 2014 May;92(3):529-42. doi: 10.1111/mmi.12571. Epub 2014 Mar 28. Mol Microbiol. 2014. PMID: 24589350 Free PMC article.
-
An rhs gene linked to the second type VI secretion cluster is a feature of the Pseudomonas aeruginosa strain PA14.J Bacteriol. 2014 Feb;196(4):800-10. doi: 10.1128/JB.00863-13. Epub 2013 Dec 6. J Bacteriol. 2014. PMID: 24317402 Free PMC article.
-
Coevolution of the ATPase ClpV, the sheath proteins TssB and TssC, and the accessory protein TagJ/HsiE1 distinguishes type VI secretion classes.J Biol Chem. 2014 Nov 21;289(47):33032-43. doi: 10.1074/jbc.M114.600510. Epub 2014 Oct 10. J Biol Chem. 2014. PMID: 25305017 Free PMC article.
References
-
- Bleves S., Viarre V., Salacha R., Michel G. P., Filloux A., Voulhoux R. (2010) Protein secretion systems in Pseudomonas aeruginosa: a wealth of pathogenic weapons. Int. J. Med. Microbiol. 300, 534–543 - PubMed
-
- Jones A. M., Dodd M. E., Morris J., Doherty C., Govan J. R., Webb A. K. (2010) Clinical outcome for cystic fibrosis patients infected with transmissible Pseudomonas aeruginosa: an 8-year prospective study. Chest 137, 1405–1409 - PubMed
-
- Filloux A., Hachani A., Bleves S. (2008) The bacterial type VI secretion machine: yet another player for protein transport across membranes. Microbiology 154, 1570–1583 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

