A survey of tools for variant analysis of next-generation genome sequencing data
- PMID: 23341494
- PMCID: PMC3956068
- DOI: 10.1093/bib/bbs086
A survey of tools for variant analysis of next-generation genome sequencing data
Abstract
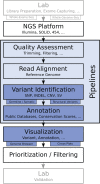
Recent advances in genome sequencing technologies provide unprecedented opportunities to characterize individual genomic landscapes and identify mutations relevant for diagnosis and therapy. Specifically, whole-exome sequencing using next-generation sequencing (NGS) technologies is gaining popularity in the human genetics community due to the moderate costs, manageable data amounts and straightforward interpretation of analysis results. While whole-exome and, in the near future, whole-genome sequencing are becoming commodities, data analysis still poses significant challenges and led to the development of a plethora of tools supporting specific parts of the analysis workflow or providing a complete solution. Here, we surveyed 205 tools for whole-genome/whole-exome sequencing data analysis supporting five distinct analytical steps: quality assessment, alignment, variant identification, variant annotation and visualization. We report an overview of the functionality, features and specific requirements of the individual tools. We then selected 32 programs for variant identification, variant annotation and visualization, which were subjected to hands-on evaluation using four data sets: one set of exome data from two patients with a rare disease for testing identification of germline mutations, two cancer data sets for testing variant callers for somatic mutations, copy number variations and structural variations, and one semi-synthetic data set for testing identification of copy number variations. Our comprehensive survey and evaluation of NGS tools provides a valuable guideline for human geneticists working on Mendelian disorders, complex diseases and cancers.
Keywords: Mendelian disorders; bioinformatics tools; cancer; next-generation sequencing; variants.
Figures
References
-
- Hodges E, Xuan Z, Balija V, et al. Genome-wide in situ exon capture for selective resequencing. Nat Genet. 2007;39:1522–7. - PubMed
-
- Rothberg JM, Hinz W, Rearick TM, et al. An integrated semiconductor device enabling non-optical genome sequencing. Nature. 2011;475:348–52. - PubMed
-
- Eisenstein M. Oxford Nanopore announcement sets sequencing sector abuzz. Nat Biotechnol. 2012;30:295–6. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

