Simplified protein design biased for prebiotic amino acids yields a foldable, halophilic protein
- PMID: 23341608
- PMCID: PMC3568330
- DOI: 10.1073/pnas.1219530110
Simplified protein design biased for prebiotic amino acids yields a foldable, halophilic protein
Abstract
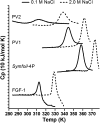
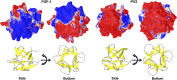
A compendium of different types of abiotic chemical syntheses identifies a consensus set of 10 "prebiotic" α-amino acids. Before the emergence of biosynthetic pathways, this set is the most plausible resource for protein formation (i.e., proteogenesis) within the overall process of abiogenesis. An essential unsolved question regarding this prebiotic set is whether it defines a "foldable set"--that is, does it contain sufficient chemical information to permit cooperatively folding polypeptides? If so, what (if any) characteristic properties might such polypeptides exhibit? To investigate these questions, two "primitive" versions of an extant protein fold (the β-trefoil) were produced by top-down symmetric deconstruction, resulting in a reduced alphabet size of 12 or 13 amino acids and a percentage of prebiotic amino acids approaching 80%. These proteins show a substantial acidification of pI and require high salt concentrations for cooperative folding. The results suggest that the prebiotic amino acids do comprise a foldable set within the halophile environment.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Reduced alphabet of prebiotic amino acids optimally encodes the conformational space of diverse extant protein folds.BMC Evol Biol. 2019 Jul 30;19(1):158. doi: 10.1186/s12862-019-1464-6. BMC Evol Biol. 2019. PMID: 31362700 Free PMC article.
-
A single aromatic core mutation converts a designed "primitive" protein from halophile to mesophile folding.Protein Sci. 2015 Jan;24(1):27-37. doi: 10.1002/pro.2580. Epub 2014 Oct 25. Protein Sci. 2015. PMID: 25297559 Free PMC article.
-
Protein design at the interface of the pre-biotic and biotic worlds.Arch Biochem Biophys. 2012 Oct 1;526(1):16-21. doi: 10.1016/j.abb.2012.06.009. Epub 2012 Jul 6. Arch Biochem Biophys. 2012. PMID: 22772066 Review.
-
Evolution and design of protein structure by folding nucleus symmetric expansion.Structure. 2014 Oct 7;22(10):1377-84. doi: 10.1016/j.str.2014.08.008. Epub 2014 Sep 18. Structure. 2014. PMID: 25242458
-
On the Evolutionary History of the Twenty Encoded Amino Acids.Chemistry. 2022 Oct 4;28(55):e202201419. doi: 10.1002/chem.202201419. Epub 2022 Jul 28. Chemistry. 2022. PMID: 35726786 Free PMC article. Review.
Cited by
-
Xeno Amino Acids: A Look into Biochemistry as We Do Not Know It.Life (Basel). 2023 Nov 29;13(12):2281. doi: 10.3390/life13122281. Life (Basel). 2023. PMID: 38137883 Free PMC article. Review.
-
Primordial emergence of a nucleic acid-binding protein via phase separation and statistical ornithine-to-arginine conversion.Proc Natl Acad Sci U S A. 2020 Jul 7;117(27):15731-15739. doi: 10.1073/pnas.2001989117. Epub 2020 Jun 19. Proc Natl Acad Sci U S A. 2020. PMID: 32561643 Free PMC article.
-
Comprehensive reduction of amino acid set in a protein suggests the importance of prebiotic amino acids for stable proteins.Sci Rep. 2018 Jan 19;8(1):1227. doi: 10.1038/s41598-018-19561-1. Sci Rep. 2018. PMID: 29352156 Free PMC article.
-
Why we are made of proteins and nucleic acids: Structural biology views on extraterrestrial life.Biophys Physicobiol. 2023 Jun 2;20(2):e200026. doi: 10.2142/biophysico.bppb-v20.0026. eCollection 2023. Biophys Physicobiol. 2023. PMID: 38496239 Free PMC article.
-
Characterization of Reconstructed Ancestral Proteins Suggests a Change in Temperature of the Ancient Biosphere.Life (Basel). 2017 Aug 6;7(3):33. doi: 10.3390/life7030033. Life (Basel). 2017. PMID: 28783077 Free PMC article. Review.
References
-
- Longo LM, Blaber M. Protein design at the interface of the pre-biotic and biotic worlds. Arch Biochem Biophys. 2012;526(1):16–21. - PubMed
-
- Doi N, Kakukawa K, Oishi Y, Yanagawa H. High solubility of random-sequence proteins consisting of five kinds of primitive amino acids. Protein Eng Des Sel. 2005;18(6):279–284. - PubMed
-
- McDonald GD, Storrie-Lombardi MC. Biochemical constraints in a protobiotic earth devoid of basic amino acids: The “BAA(-) world.”. Astrobiology. 2010;10(10):989–1000. - PubMed
-
- Huber C, Wächtershäuser G. α-Hydroxy and α-amino acids under possible Hadean, volcanic origin-of-life conditions. Science. 2006;314(5799):630–632. - PubMed
-
- Lerner NR, Peterson E, Chang S. The Strecker synthesis as a source of amino acids in carbonaceous chondrites: Deuterium retention during synthesis. Geochim Cosmochim Acta. 1993;57(19):4713–4723. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

