Aberrant splicing of HTT generates the pathogenic exon 1 protein in Huntington disease
- PMID: 23341618
- PMCID: PMC3568346
- DOI: 10.1073/pnas.1221891110
Aberrant splicing of HTT generates the pathogenic exon 1 protein in Huntington disease
Abstract
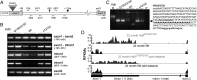
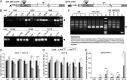
Huntington disease (HD) is a devastating, late-onset, inherited neurodegenerative disorder that manifests with personality changes, movement disorders, and cognitive decline. It is caused by a CAG repeat expansion in exon 1 of the HTT gene that translates to a polyglutamine tract in the huntingtin protein (HTT). The formation of HTT fragments has been implicated as an essential step in the molecular pathogenesis of HD and several proteases that cleave HTT have been identified. However, the importance of smaller N-terminal fragments has been highlighted by their presence in HD postmortem brains and by the fact that nuclear inclusions are only detected by antibodies to the N terminus of HTT. Despite an intense research effort, the precise length of these fragments and the mechanism by which they are generated remains unknown. Here we show that CAG repeat length-dependent aberrant splicing of exon 1 HTT results in a short polyadenylated mRNA that is translated into an exon 1 HTT protein. Given that mutant exon 1 HTT proteins have consistently been shown to be highly pathogenic in HD mouse models, the aberrant splicing of HTT mRNA provides a mechanistic basis for the molecular pathogenesis of HD. RNA-targeted therapeutic strategies designed to lower the levels of HTT are under development. Many of these approaches would not prevent the production of exon 1 HTT and should be reviewed in light of our findings.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Comment in
-
Neurodegenerative disease: altered DNA methylation and RNA splicing could be key mechanisms in Huntington disease.Nat Rev Neurol. 2013 Mar;9(3):119. doi: 10.1038/nrneurol.2013.23. Epub 2013 Feb 12. Nat Rev Neurol. 2013. PMID: 23399643 No abstract available.
-
Aberrantly spliced HTT, a new player in Huntington's disease pathogenesis.RNA Biol. 2013 Nov;10(11):1647-52. doi: 10.4161/rna.26706. Epub 2013 Oct 11. RNA Biol. 2013. PMID: 24256709 Free PMC article.
References
-
- Bates GP, Harper PS, Jones AL, eds (2002) Huntington's Disease (Oxford University Press, Oxford), 3rd Ed.
-
- Ross CA, Tabrizi SJ. Huntington’s disease: From molecular pathogenesis to clinical treatment. Lancet Neurol. 2011;10(1):83–98. - PubMed
-
- Gafni J, et al. Inhibition of calpain cleavage of huntingtin reduces toxicity: Accumulation of calpain/caspase fragments in the nucleus. J Biol Chem. 2004;279(19):20211–20220. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

