Evolutionary optimization of protein folding
- PMID: 23341762
- PMCID: PMC3547816
- DOI: 10.1371/journal.pcbi.1002861
Evolutionary optimization of protein folding
Abstract
Nature has shaped the make up of proteins since their appearance, [Formula: see text]3.8 billion years ago. However, the fundamental drivers of structural change responsible for the extraordinary diversity of proteins have yet to be elucidated. Here we explore if protein evolution affects folding speed. We estimated folding times for the present-day catalog of protein domains directly from their size-modified contact order. These values were mapped onto an evolutionary timeline of domain appearance derived from a phylogenomic analysis of protein domains in 989 fully-sequenced genomes. Our results show a clear overall increase of folding speed during evolution, with known ultra-fast downhill folders appearing rather late in the timeline. Remarkably, folding optimization depends on secondary structure. While alpha-folds showed a tendency to fold faster throughout evolution, beta-folds exhibited a trend of folding time increase during the last [Formula: see text]1.5 billion years that began during the "big bang" of domain combinations. As a consequence, these domain structures are on average slow folders today. Our results suggest that fast and efficient folding of domains shaped the universe of protein structure. This finding supports the hypothesis that optimization of the kinetic and thermodynamic accessibility of the native fold reduces protein aggregation propensities that hamper cellular functions.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

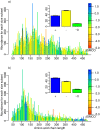
 SMCO, the difference between the end points of the polynomial regression of SMCO in this dataset, for the specified initial (a) and later (b) time period. Yellow to red indicates a decrease, and blue an increase in SMCO. The barplots (inset) show the percentage of domains with positive (blue), negative (yellow), and insignificant (green)
SMCO, the difference between the end points of the polynomial regression of SMCO in this dataset, for the specified initial (a) and later (b) time period. Yellow to red indicates a decrease, and blue an increase in SMCO. The barplots (inset) show the percentage of domains with positive (blue), negative (yellow), and insignificant (green)  SMCO.
SMCO.
 1.5 Gya, and b)
1.5 Gya, and b)  1.5-0 Gya. Each barplot considers one of the four fold classes according to their secondary structure: all-
1.5-0 Gya. Each barplot considers one of the four fold classes according to their secondary structure: all- , all-
, all- ,
,  /
/ , and
, and  +
+ , as indicated. The barplots were obtained from domain length distributions analogous to those shown in Figure S3.
, as indicated. The barplots were obtained from domain length distributions analogous to those shown in Figure S3.Similar articles
-
Tracing protein and proteome history with chronologies and networks: folding recapitulates evolution.Expert Rev Proteomics. 2021 Oct;18(10):863-880. doi: 10.1080/14789450.2021.1992277. Epub 2021 Nov 22. Expert Rev Proteomics. 2021. PMID: 34628994 Review.
-
Factors governing the foldability of proteins.Proteins. 1996 Dec;26(4):411-41. doi: 10.1002/(SICI)1097-0134(199612)26:4<411::AID-PROT4>3.0.CO;2-E. Proteins. 1996. PMID: 8990496
-
Proteome evolution and the metabolic origins of translation and cellular life.J Mol Evol. 2011 Jan;72(1):14-33. doi: 10.1007/s00239-010-9400-9. Epub 2010 Nov 17. J Mol Evol. 2011. PMID: 21082171
-
Limited cooperativity in protein folding.Curr Opin Struct Biol. 2016 Feb;36:58-66. doi: 10.1016/j.sbi.2015.12.001. Epub 2016 Feb 2. Curr Opin Struct Biol. 2016. PMID: 26845039 Review.
-
The folding landscape of an alpha-lytic protease variant reveals the role of a conserved beta-hairpin in the development of kinetic stability.Proteins. 2005 Oct 1;61(1):105-14. doi: 10.1002/prot.20525. Proteins. 2005. PMID: 16044461
Cited by
-
Evolution of networks of protein domain organization.Sci Rep. 2021 Jun 8;11(1):12075. doi: 10.1038/s41598-021-90498-8. Sci Rep. 2021. PMID: 34103558 Free PMC article.
-
The organization of domains in proteins obeys Menzerath-Altmann's law of language.BMC Syst Biol. 2015 Aug 11;9:44. doi: 10.1186/s12918-015-0192-9. BMC Syst Biol. 2015. PMID: 26260760 Free PMC article.
-
Selective pressure for rapid membrane integration constrains the sequence of bacterial outer membrane proteins.Mol Microbiol. 2017 Dec;106(5):777-792. doi: 10.1111/mmi.13845. Epub 2017 Oct 16. Mol Microbiol. 2017. PMID: 28941249 Free PMC article.
-
Folding Rate Optimization Promotes Frustrated Interactions in Entangled Protein Structures.Int J Mol Sci. 2019 Dec 27;21(1):213. doi: 10.3390/ijms21010213. Int J Mol Sci. 2019. PMID: 31892272 Free PMC article.
-
Evolutionary trend toward kinetic stability in the folding trajectory of RNases H.Proc Natl Acad Sci U S A. 2016 Nov 15;113(46):13045-13050. doi: 10.1073/pnas.1611781113. Epub 2016 Oct 31. Proc Natl Acad Sci U S A. 2016. PMID: 27799545 Free PMC article.
References
-
- Qiu L, Pabit SA, Roitberg AE, Hagen SJ (2002) Smaller and faster: the 20-residue trp-cage protein folds in 4 micros. J Am Chem Soc 124: 12952–12953. - PubMed
-
- Goldberg ME, Semisotnov GV, Friguet B, Kuwajima K, Ptitsyn OB, et al. (1990) An early immunoreactive folding intermediate of the tryptophan synthase 2 subunit is a molten globule. FEBS Letters 263: 51–56. - PubMed
-
- Matagne A, Chung EW, Ball LJ, Radford SE, Robinson CV, et al. (1998) The origin of the alphadomain intermediate in the folding of hen lysozyme. J Mol Biol 277: 997–1005. - PubMed
-
- Onuchic JN, Wolynes PG (2004) Theory of protein folding. Curr Opin Struct Biol 14: 70–75. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

