Binding of nucleoid-associated protein fis to DNA is regulated by DNA breathing dynamics
- PMID: 23341768
- PMCID: PMC3547798
- DOI: 10.1371/journal.pcbi.1002881
Binding of nucleoid-associated protein fis to DNA is regulated by DNA breathing dynamics
Abstract
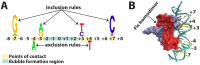
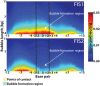
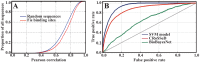
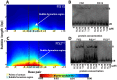
Physicochemical properties of DNA, such as shape, affect protein-DNA recognition. However, the properties of DNA that are most relevant for predicting the binding sites of particular transcription factors (TFs) or classes of TFs have yet to be fully understood. Here, using a model that accurately captures the melting behavior and breathing dynamics (spontaneous local openings of the double helix) of double-stranded DNA, we simulated the dynamics of known binding sites of the TF and nucleoid-associated protein Fis in Escherichia coli. Our study involves simulations of breathing dynamics, analysis of large published in vitro and genomic datasets, and targeted experimental tests of our predictions. Our simulation results and available in vitro binding data indicate a strong correlation between DNA breathing dynamics and Fis binding. Indeed, we can define an average DNA breathing profile that is characteristic of Fis binding sites. This profile is significantly enriched among the identified in vivo E. coli Fis binding sites. To test our understanding of how Fis binding is influenced by DNA breathing dynamics, we designed base-pair substitutions, mismatch, and methylation modifications of DNA regions that are known to interact (or not interact) with Fis. The goal in each case was to make the local DNA breathing dynamics either closer to or farther from the breathing profile characteristic of a strong Fis binding site. For the modified DNA segments, we found that Fis-DNA binding, as assessed by gel-shift assay, changed in accordance with our expectations. We conclude that Fis binding is associated with DNA breathing dynamics, which in turn may be regulated by various nucleotide modifications.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

Similar articles
-
Biochemical identification of base and phosphate contacts between Fis and a high-affinity DNA binding site.J Mol Biol. 2008 Jul 4;380(2):327-39. doi: 10.1016/j.jmb.2008.04.075. Epub 2008 May 7. J Mol Biol. 2008. PMID: 18514225 Free PMC article.
-
Common and variable contributions of Fis residues to high-affinity binding at different DNA sequences.J Bacteriol. 2006 Mar;188(6):2081-95. doi: 10.1128/JB.188.6.2081-2095.2006. J Bacteriol. 2006. PMID: 16513738 Free PMC article.
-
Information analysis of Fis binding sites.Nucleic Acids Res. 1997 Dec 15;25(24):4994-5002. doi: 10.1093/nar/25.24.4994. Nucleic Acids Res. 1997. PMID: 9396807 Free PMC article.
-
Multiple Binding Configurations of Fis Protein Pairs on DNA: Facilitated Dissociation versus Cooperative Dissociation.J Am Chem Soc. 2019 Nov 13;141(45):18113-18126. doi: 10.1021/jacs.9b08287. Epub 2019 Nov 1. J Am Chem Soc. 2019. PMID: 31566963
-
DNA supercoiling and transcription in Escherichia coli: The FIS connection.Biochimie. 2001 Feb;83(2):213-7. doi: 10.1016/s0300-9084(00)01217-7. Biochimie. 2001. PMID: 11278071 Review.
Cited by
-
Analysis of the leakage of gene repression by an artificial TetR-regulated promoter in cyanobacteria.BMC Res Notes. 2015 Sep 19;8:459. doi: 10.1186/s13104-015-1425-0. BMC Res Notes. 2015. PMID: 26387086 Free PMC article.
-
Examining DNA breathing with pyDNA-EPBD.Bioinformatics. 2023 Nov 1;39(11):btad699. doi: 10.1093/bioinformatics/btad699. Bioinformatics. 2023. PMID: 37991847 Free PMC article.
-
Mesoscopic model and free energy landscape for protein-DNA binding sites: analysis of cyanobacterial promoters.PLoS Comput Biol. 2014 Oct 2;10(10):e1003835. doi: 10.1371/journal.pcbi.1003835. eCollection 2014 Oct. PLoS Comput Biol. 2014. PMID: 25275384 Free PMC article.
-
Propionylation of Fis K32 in Salmonella enterica serovar Typhi: a key modification affecting pathogenicity.Future Microbiol. 2025 Mar;20(4):295-303. doi: 10.1080/17460913.2025.2460338. Epub 2025 Jan 30. Future Microbiol. 2025. PMID: 39885648
-
Evaluating the role of coherent delocalized phonon-like modes in DNA cyclization.Sci Rep. 2017 Aug 29;7(1):9731. doi: 10.1038/s41598-017-09537-y. Sci Rep. 2017. PMID: 28851939 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

