Atomic model of rabbit hemorrhagic disease virus by cryo-electron microscopy and crystallography
- PMID: 23341770
- PMCID: PMC3547835
- DOI: 10.1371/journal.ppat.1003132
Atomic model of rabbit hemorrhagic disease virus by cryo-electron microscopy and crystallography
Abstract
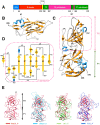
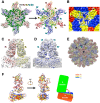
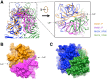
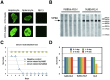
Rabbit hemorrhagic disease, first described in China in 1984, causes hemorrhagic necrosis of the liver. Its etiological agent, rabbit hemorrhagic disease virus (RHDV), belongs to the Lagovirus genus in the family Caliciviridae. The detailed molecular structure of any lagovirus capsid has yet to be determined. Here, we report a cryo-electron microscopic (cryoEM) reconstruction of wild-type RHDV at 6.5 Å resolution and the crystal structures of the shell (S) and protruding (P) domains of its major capsid protein, VP60, each at 2.0 Å resolution. From these data we built a complete atomic model of the RHDV capsid. VP60 has a conserved S domain and a specific P2 sub-domain that differs from those found in other caliciviruses. As seen in the shell portion of the RHDV cryoEM map, which was resolved to ~5.5 Å, the N-terminal arm domain of VP60 folds back onto its cognate S domain. Sequence alignments of VP60 from six groups of RHDV isolates revealed seven regions of high variation that could be mapped onto the surface of the P2 sub-domain and suggested three putative pockets might be responsible for binding to histo-blood group antigens. A flexible loop in one of these regions was shown to interact with rabbit tissue cells and contains an important epitope for anti-RHDV antibody production. Our study provides a reliable, pseudo-atomic model of a Lagovirus and suggests a new candidate for an efficient vaccine that can be used to protect rabbits from RHDV infection.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


Similar articles
-
Cryo-electron microscopy reconstructions of two types of wild rabbit hemorrhagic disease viruses characterized the structural features of Lagovirus.Protein Cell. 2010 Jan;1(1):48-58. doi: 10.1007/s13238-010-0007-0. Epub 2010 Feb 7. Protein Cell. 2010. PMID: 21203997 Free PMC article.
-
Structural Basis for Rabbit Hemorrhagic Disease Virus Antibody Specificity.J Virol. 2022 Nov 23;96(22):e0121722. doi: 10.1128/jvi.01217-22. Epub 2022 Nov 3. J Virol. 2022. PMID: 36326275 Free PMC article.
-
Egg yolk IgY against RHDV capsid protein VP60 promotes rabbit defense against RHDV infection.Vet Immunol Immunopathol. 2014 Jan 15;157(1-2):97-104. doi: 10.1016/j.vetimm.2013.10.002. Epub 2013 Oct 30. Vet Immunol Immunopathol. 2014. PMID: 24252247
-
Genetic variation and phylogenetic analysis of rabbit haemorrhagic disease virus (RHDV) strains.Acta Biochim Pol. 2012;59(4):459-65. Epub 2012 Dec 13. Acta Biochim Pol. 2012. PMID: 23240105 Review.
-
Lack of evidence for differences in the spread of classic (Lagovirus europaeus/GI.1) and novel (Lagovirus europaeus/GI.2) rabbit haemorrhagic disease viruses in Europe and North Africa.Vet Rec. 2022 Feb;190(3):e1067. doi: 10.1002/vetr.1067. Epub 2021 Oct 28. Vet Rec. 2022. PMID: 34713453 Review.
Cited by
-
Biological cryo-electron microscopy in China.Protein Sci. 2017 Jan;26(1):16-31. doi: 10.1002/pro.3018. Epub 2016 Sep 2. Protein Sci. 2017. PMID: 27534377 Free PMC article. Review.
-
Rabbit hemorrhagic disease virus capsid, a versatile platform for foreign B-cell epitope display inducing protective humoral immune responses.Sci Rep. 2016 Aug 23;6:31844. doi: 10.1038/srep31844. Sci Rep. 2016. PMID: 27549017 Free PMC article.
-
Chimeric RHDV Virus-Like Particles Displaying Foot-and-Mouth Disease Virus Epitopes Elicit Neutralizing Antibodies and Confer Partial Protection in Pigs.Vaccines (Basel). 2021 May 7;9(5):470. doi: 10.3390/vaccines9050470. Vaccines (Basel). 2021. PMID: 34066934 Free PMC article.
-
Recombination between non-structural and structural genes as a mechanism of selection in lagoviruses: The evolutionary dead-end of an RHDV2 isolated from European hare.Virus Res. 2024 Jan 2;339:199257. doi: 10.1016/j.virusres.2023.199257. Epub 2023 Nov 3. Virus Res. 2024. PMID: 38347757 Free PMC article.
-
Precise location of linear epitopes on the capsid surface of feline calicivirus recognized by neutralizing and non-neutralizing monoclonal antibodies.Vet Res. 2020 May 1;51(1):59. doi: 10.1186/s13567-020-00785-x. Vet Res. 2020. PMID: 32357948 Free PMC article.
References
-
- Xu ZJ, Chen WX (1989) Viral haemorrhagic disease in rabbits:a review. Vet Res Commun 13: 205–212. - PubMed
-
- Liu SJ, Xue HP, Pu BQ, Qian NH (1984) A new viral disease in rabbits. AnimHusbVet Med 16: 253–255.
-
- Nowotny N, Bascunana CR, Ballagi-Pordany A, Gavier-Widen D, Uhlen M, et al. (1997) Phylogenetic analysis of rabbit haemorrhagic disease and European brown hare syndrome viruses by comparison of sequences from the capsid protein gene. Arch Virol 142: 657–673. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

