A systematic mapping approach of 16q12.2/FTO and BMI in more than 20,000 African Americans narrows in on the underlying functional variation: results from the Population Architecture using Genomics and Epidemiology (PAGE) study
- PMID: 23341774
- PMCID: PMC3547789
- DOI: 10.1371/journal.pgen.1003171
A systematic mapping approach of 16q12.2/FTO and BMI in more than 20,000 African Americans narrows in on the underlying functional variation: results from the Population Architecture using Genomics and Epidemiology (PAGE) study
Abstract
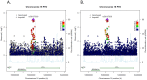
Genetic variants in intron 1 of the fat mass- and obesity-associated (FTO) gene have been consistently associated with body mass index (BMI) in Europeans. However, follow-up studies in African Americans (AA) have shown no support for some of the most consistently BMI-associated FTO index single nucleotide polymorphisms (SNPs). This is most likely explained by different race-specific linkage disequilibrium (LD) patterns and lower correlation overall in AA, which provides the opportunity to fine-map this region and narrow in on the functional variant. To comprehensively explore the 16q12.2/FTO locus and to search for second independent signals in the broader region, we fine-mapped a 646-kb region, encompassing the large FTO gene and the flanking gene RPGRIP1L by investigating a total of 3,756 variants (1,529 genotyped and 2,227 imputed variants) in 20,488 AAs across five studies. We observed associations between BMI and variants in the known FTO intron 1 locus: the SNP with the most significant p-value, rs56137030 (8.3 × 10(-6)) had not been highlighted in previous studies. While rs56137030was correlated at r(2)>0.5 with 103 SNPs in Europeans (including the GWAS index SNPs), this number was reduced to 28 SNPs in AA. Among rs56137030 and the 28 correlated SNPs, six were located within candidate intronic regulatory elements, including rs1421085, for which we predicted allele-specific binding affinity for the transcription factor CUX1, which has recently been implicated in the regulation of FTO. We did not find strong evidence for a second independent signal in the broader region. In summary, this large fine-mapping study in AA has substantially reduced the number of common alleles that are likely to be functional candidates of the known FTO locus. Importantly our study demonstrated that comprehensive fine-mapping in AA provides a powerful approach to narrow in on the functional candidate(s) underlying the initial GWAS findings in European populations.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
References
-
- Thorleifsson G, Walters GB, Gudbjartsson DF, Steinthorsdottir V, Sulem P, et al. (2009) Genome-wide association yields new sequence variants at seven loci that associate with measures of obesity. Nat Genet 41: 18–24. - PubMed
-
- Hinney A, Nguyen TT, Scherag A, Friedel S, Bronner G, et al. (2007) Genome wide association (GWA) study for early onset extreme obesity supports the role of fat mass and obesity associated gene (FTO) variants. PLoS ONE 2: e1361 doi:10.1371/journal.pone.0001361. - DOI - PMC - PubMed
-
- Scuteri A, Sanna S, Chen WM, Uda M, Albai G, et al. (2007) Genome-wide association scan shows genetic variants in the FTO gene are associated with obesity-related traits. PLoS Genet 3: e115 doi:10.1371/journal.pgen.0030115. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- U01 HG004790/HG/NHGRI NIH HHS/United States
- N01-HC-55016/HC/NHLBI NIH HHS/United States
- 24152/PHS HHS/United States
- N01 HC055020/HL/NHLBI NIH HHS/United States
- N01 HC055018/HL/NHLBI NIH HHS/United States
- U01HG004801/HG/NHGRI NIH HHS/United States
- N01-HC-55018/HC/NHLBI NIH HHS/United States
- P01CA33619/CA/NCI NIH HHS/United States
- 32105-6/PHS HHS/United States
- N01 HC055016/HL/NHLBI NIH HHS/United States
- U01CA98758/CA/NCI NIH HHS/United States
- N01 HC055019/HL/NHLBI NIH HHS/United States
- N01 HC055021/HL/NHLBI NIH HHS/United States
- N01-HC-55022/HC/NHLBI NIH HHS/United States
- U01 HG004802/HG/NHGRI NIH HHS/United States
- 42107-26/PHS HHS/United States
- 32115/PHS HHS/United States
- N01 WH022110/WH/WHI NIH HHS/United States
- U01CA136792/CA/NCI NIH HHS/United States
- N01 HC055015/HL/NHLBI NIH HHS/United States
- U01HG004803/HG/NHGRI NIH HHS/United States
- K99 DK091318/DK/NIDDK NIH HHS/United States
- N01-HC-55021/HC/NHLBI NIH HHS/United States
- U01HG004802/HG/NHGRI NIH HHS/United States
- 32100-2/PHS HHS/United States
- 42129-32/PHS HHS/United States
- N01-HC-55019/HC/NHLBI NIH HHS/United States
- N01-HC-55015/HC/NHLBI NIH HHS/United States
- U01 HG004803/HG/NHGRI NIH HHS/United States
- 1K99DK091318-01/DK/NIDDK NIH HHS/United States
- 32118-32119/PHS HHS/United States
- P01 CA033619/CA/NCI NIH HHS/United States
- U01HG004798/HG/NHGRI NIH HHS/United States
- N01-HC-55020/HC/NHLBI NIH HHS/United States
- 32108-9/PHS HHS/United States
- R37CA54281/CA/NCI NIH HHS/United States
- U01 CA098758/CA/NCI NIH HHS/United States
- N01 HC055022/HL/NHLBI NIH HHS/United States
- U01HG004790/HG/NHGRI NIH HHS/United States
- U01 HG004798/HG/NHGRI NIH HHS/United States
- 32122/PHS HHS/United States
- 44221/PHS HHS/United States
- U01 CA136792/CA/NCI NIH HHS/United States
- 32111-13/PHS HHS/United States
- R37 CA054281/CA/NCI NIH HHS/United States
- R01 CA63/CA/NCI NIH HHS/United States
- U01 HG004801/HG/NHGRI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

