Multiple opposing constraints govern chromosome interactions during meiosis
- PMID: 23341780
- PMCID: PMC3547833
- DOI: 10.1371/journal.pgen.1003197
Multiple opposing constraints govern chromosome interactions during meiosis
Abstract
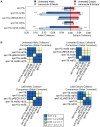
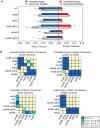
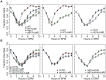
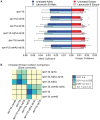
Homolog pairing and crossing over during meiosis I prophase is required for accurate chromosome segregation to form euploid gametes. The repair of Spo11-induced double-strand breaks (DSB) using a homologous chromosome template is a major driver of pairing in many species, including fungi, plants, and mammals. Inappropriate pairing and crossing over at ectopic loci can lead to chromosome rearrangements and aneuploidy. How (or if) inappropriate ectopic interactions are disrupted in favor of allelic interactions is not clear. Here we used an in vivo "collision" assay in budding yeast to test the contributions of cohesion and the organization and motion of chromosomes in the nucleus on promoting or antagonizing interactions between allelic and ectopic loci at interstitial chromosome sites. We found that deletion of the cohesin subunit Rec8, but not other chromosome axis proteins (e.g. Red1, Hop1, or Mek1), caused an increase in homolog-nonspecific chromosome interaction, even in the absence of Spo11. This effect was partially suppressed by expression of the mitotic cohesin paralog Scc1/Mdc1, implicating Rec8's role in cohesion rather than axis integrity in preventing nonspecific chromosome interactions. Disruption of telomere-led motion by treating cells with the actin polymerization inhibitor Latrunculin B (Lat B) elevated nonspecific collisions in rec8Δ spo11Δ. Next, using a visual homolog-pairing assay, we found that the delay in homolog pairing in mutants defective for telomere-led chromosome motion (ndj1Δ or csm4Δ) is enhanced in Lat B-treated cells, implicating actin in more than one process promoting homolog juxtaposition. We suggest that multiple, independent contributions of actin, cohesin, and telomere function are integrated to promote stable homolog-specific interactions and to destabilize weak nonspecific interactions by modulating the elastic spring-like properties of chromosomes.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



Similar articles
-
Pds5 is required for homologue pairing and inhibits synapsis of sister chromatids during yeast meiosis.J Cell Biol. 2009 Sep 7;186(5):713-25. doi: 10.1083/jcb.200810107. J Cell Biol. 2009. PMID: 19736318 Free PMC article.
-
Meiotic chromosome pairing is promoted by telomere-led chromosome movements independent of bouquet formation.PLoS Genet. 2012;8(5):e1002730. doi: 10.1371/journal.pgen.1002730. Epub 2012 May 24. PLoS Genet. 2012. PMID: 22654677 Free PMC article.
-
How do small chromosomes know they are small? Maximizing meiotic break formation on the shortest yeast chromosomes.Curr Genet. 2021 Jun;67(3):431-437. doi: 10.1007/s00294-021-01160-9. Epub 2021 Feb 18. Curr Genet. 2021. PMID: 33604699 Free PMC article. Review.
-
Meiotic cohesin SMC1β provides prophase I centromeric cohesion and is required for multiple synapsis-associated functions.PLoS Genet. 2013;9(12):e1003985. doi: 10.1371/journal.pgen.1003985. Epub 2013 Dec 26. PLoS Genet. 2013. PMID: 24385917 Free PMC article.
-
Centromere pairing precedes meiotic chromosome pairing in plants.Sci China Life Sci. 2017 Nov;60(11):1197-1202. doi: 10.1007/s11427-017-9109-y. Epub 2017 Jul 26. Sci China Life Sci. 2017. PMID: 28755295 Review.
Cited by
-
Modeling meiotic chromosome pairing: nuclear envelope attachment, telomere-led active random motion, and anomalous diffusion.Phys Biol. 2016 Apr 5;13(2):026003. doi: 10.1088/1478-3975/13/2/026003. Phys Biol. 2016. PMID: 27046097 Free PMC article.
-
Modeling cell biological features of meiotic chromosome pairing to study interlock resolution.PLoS Comput Biol. 2022 Jun 13;18(6):e1010252. doi: 10.1371/journal.pcbi.1010252. eCollection 2022 Jun. PLoS Comput Biol. 2022. PMID: 35696428 Free PMC article.
-
Diffusion and distal linkages govern interchromosomal dynamics during meiotic prophase.Proc Natl Acad Sci U S A. 2022 Mar 22;119(12):e2115883119. doi: 10.1073/pnas.2115883119. Epub 2022 Mar 18. Proc Natl Acad Sci U S A. 2022. PMID: 35302885 Free PMC article.
-
The Nucleoporin Nup2 Contains a Meiotic-Autonomous Region that Promotes the Dynamic Chromosome Events of Meiosis.Genetics. 2017 Jul;206(3):1319-1337. doi: 10.1534/genetics.116.194555. Epub 2017 Apr 28. Genetics. 2017. PMID: 28455351 Free PMC article.
-
The Chromosomal Courtship Dance-homolog pairing in early meiosis.Curr Opin Cell Biol. 2014 Feb;26:123-31. doi: 10.1016/j.ceb.2013.12.004. Epub 2014 Jan 7. Curr Opin Cell Biol. 2014. PMID: 24529254 Free PMC article. Review.
References
-
- Gerton JL, Hawley RS (2005) Homologous chromosome interactions in meiosis: diversity amidst conservation. Nat Rev Gen 6: 477–487. - PubMed
-
- Tsai JH, McKee BD (2011) Homologous pairing and the role of pairing centers in meiosis. J Cell Sci 124: 1955–1963. - PubMed
-
- Hunt PA, Hassold TJ (2008) Human female meiosis: What makes a good egg go bad? Trends Genet 24: 86–93. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

