Human disease-associated genetic variation impacts large intergenic non-coding RNA expression
- PMID: 23341781
- PMCID: PMC3547830
- DOI: 10.1371/journal.pgen.1003201
Human disease-associated genetic variation impacts large intergenic non-coding RNA expression
Abstract
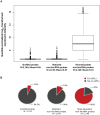
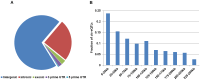
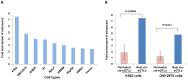
Recently it has become clear that only a small percentage (7%) of disease-associated single nucleotide polymorphisms (SNPs) are located in protein-coding regions, while the remaining 93% are located in gene regulatory regions or in intergenic regions. Thus, the understanding of how genetic variations control the expression of non-coding RNAs (in a tissue-dependent manner) has far-reaching implications. We tested the association of SNPs with expression levels (eQTLs) of large intergenic non-coding RNAs (lincRNAs), using genome-wide gene expression and genotype data from five different tissues. We identified 112 cis-regulated lincRNAs, of which 45% could be replicated in an independent dataset. We observed that 75% of the SNPs affecting lincRNA expression (lincRNA cis-eQTLs) were specific to lincRNA alone and did not affect the expression of neighboring protein-coding genes. We show that this specific genotype-lincRNA expression correlation is tissue-dependent and that many of these lincRNA cis-eQTL SNPs are also associated with complex traits and diseases.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
Similar articles
-
Genetic and epigenetic regulation of human lincRNA gene expression.Am J Hum Genet. 2013 Dec 5;93(6):1015-26. doi: 10.1016/j.ajhg.2013.10.022. Epub 2013 Nov 21. Am J Hum Genet. 2013. PMID: 24268656 Free PMC article.
-
Uncovering association networks through an eQTL analysis involving human miRNAs and lincRNAs.Sci Rep. 2018 Oct 9;8(1):15050. doi: 10.1038/s41598-018-33420-z. Sci Rep. 2018. PMID: 30301969 Free PMC article.
-
cis-Acting Complex-Trait-Associated lincRNA Expression Correlates with Modulation of Chromosomal Architecture.Cell Rep. 2017 Feb 28;18(9):2280-2288. doi: 10.1016/j.celrep.2017.02.009. Cell Rep. 2017. PMID: 28249171
-
Expression Quantitative Trait Loci Information Improves Predictive Modeling of Disease Relevance of Non-Coding Genetic Variation.PLoS One. 2015 Oct 16;10(10):e0140758. doi: 10.1371/journal.pone.0140758. eCollection 2015. PLoS One. 2015. PMID: 26474488 Free PMC article. Review.
-
Significance of Single-Nucleotide Variants in Long Intergenic Non-protein Coding RNAs.Front Cell Dev Biol. 2020 May 25;8:347. doi: 10.3389/fcell.2020.00347. eCollection 2020. Front Cell Dev Biol. 2020. PMID: 32523949 Free PMC article. Review.
Cited by
-
Dissecting the genetics of the human transcriptome identifies novel trait-related trans-eQTLs and corroborates the regulatory relevance of non-protein coding loci†.Hum Mol Genet. 2015 Aug 15;24(16):4746-63. doi: 10.1093/hmg/ddv194. Epub 2015 May 27. Hum Mol Genet. 2015. PMID: 26019233 Free PMC article. Clinical Trial.
-
Methods for the Analysis and Interpretation for Rare Variants Associated with Complex Traits.Curr Protoc Hum Genet. 2019 Apr;101(1):e83. doi: 10.1002/cphg.83. Epub 2019 Mar 8. Curr Protoc Hum Genet. 2019. PMID: 30849219 Free PMC article. Review.
-
Negative Association Between lncRNA HOTTIP rs3807598 C>G and Hirschsprung Disease.Pharmgenomics Pers Med. 2020 May 6;13:151-156. doi: 10.2147/PGPM.S249649. eCollection 2020. Pharmgenomics Pers Med. 2020. PMID: 32440194 Free PMC article.
-
Genome-wide identification and functional prediction of novel and drought-responsive lincRNAs in Populus trichocarpa.J Exp Bot. 2014 Sep;65(17):4975-83. doi: 10.1093/jxb/eru256. Epub 2014 Jun 19. J Exp Bot. 2014. PMID: 24948679 Free PMC article.
-
The Role of lncRNA in the Development of Tumors, including Breast Cancer.Int J Mol Sci. 2021 Aug 5;22(16):8427. doi: 10.3390/ijms22168427. Int J Mol Sci. 2021. PMID: 34445129 Free PMC article. Review.
References
-
- Kapranov P, Cheng J, Dike S, Nix DA, Duttagupta R, et al. (2007) RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science 316: 1484–1488. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

