The HPV E2-Host Protein-Protein Interactions: A Complex Hijacking of the Cellular Network
- PMID: 23341853
- PMCID: PMC3547520
- DOI: 10.2174/1874357901206010173
The HPV E2-Host Protein-Protein Interactions: A Complex Hijacking of the Cellular Network
Abstract
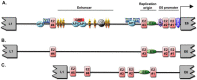
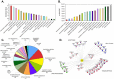
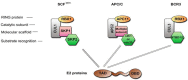
Over 100 genotypes of human papillomaviruses (HPVs) have been identified as being responsible for unapparent infections or for lesions ranging from benign skin or genital warts to cancer. The pathogenesis of HPV results from complex relationships between viral and host factors, driven in particular by the interplay between the host proteome and the early viral proteins. The E2 protein regulates the transcription, the replication as well as the mitotic segregation of the viral genome through the recruitment of host cell factors to the HPV regulatory region. It is thereby a pivotal factor for the productive viral life cycle and for viral persistence, a major risk factor for cancer development. In addition, the E2 proteins have been shown to engage numerous interactions through which they play important roles in modulating the host cell. Such E2 activities are probably contributing to create cell conditions appropriate for the successive stages of the viral life cycle, and some of these activities have been demonstrated only for the oncogenic high-risk HPV. The recent mapping of E2-host protein-protein interactions with 12 genotypes representative of HPV diversity has shed some light on the large complexity of the host cell hijacking and on its diversity according to viral genotypes. This article reviews the functions of E2 as they emerge from the E2/host proteome interplay, taking into account the large-scale comparative interactomic study.
Keywords: E2; HPV; cervical cancer.; network; productive life cycle; protein interactions; viral pathogenesis.
Figures


References
-
- Hegde RS, Androphy EJ. Crystal structure of the E2 DNA-binding domain from human papillomavirus type 16: implications for its DNA binding-site selection mechanism. J Mol Biol. 1998;284:1479–89. - PubMed
-
- Antson AA, Burns JE, Moroz OV, et al. Structure of the intact transactivation domain of the human papillomavirus E2 protein. Nature. 2000;403(6771):805–9. - PubMed
-
- Bochkarev A, Barwell JA, Pfuetzner RA, Bochkareva E, Frappier L, Edwards AM. Crystal structure of the DNA-binding domain of the Epstein-Barr virus origin-binding protein, EBNA1, bound to DNA. Cell. 1996;84:791–800. - PubMed
-
- McBride AA, Oliveira JG, McPhillips MG. Partitioning viral genomes in mitosis: same idea, different targets. Cell Cycle. 2006;5:1499–502. - PubMed
-
- Boulabiar M, Boubaker S, Favre M, Demeret C. Keratinocyte sensitization to tumour necrosis factor-induced nuclear factor kappa B activation by the E2 regulatory protein of human papillomaviruses. J Gen Virol. 2011;92:2422–7. - PubMed
LinkOut - more resources
Full Text Sources
