Identification of upregulated genes under cold stress in cold-tolerant chickpea using the cDNA-AFLP approach
- PMID: 23341906
- PMCID: PMC3544839
- DOI: 10.1371/journal.pone.0052757
Identification of upregulated genes under cold stress in cold-tolerant chickpea using the cDNA-AFLP approach
Abstract
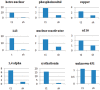
Low temperature injury is one of the most significant causes of crop damage worldwide. Cold acclimatization processes improve the freezing tolerance of plants. To identify genes of potential importance for acclimatzation to the cold and to elucidate the pathways that regulate this process, global transcriptome expression of the chickpea (Cicer arietinum L), a species of legume, was analyzed using the cDNA-AFLP technique. In total, we generated 4800 transcript-derived fragments (TDFs) using cDNA-AFLP in conjunction with 256 primer combinations. We only considered those cDNA fragments that seemed to be up-regulated during cold acclimatization. Of these, 102 TDFs with differential expression patterns were excised from gels and re-amplified by PCR. Fifty-four fragments were then cloned and sequenced. BLAST search of the GenBank non-redundant (nr) sequence database demonstrated that 77 percent of the TDFs belonged to known sequences with putative functions related to metabolism (31), transport (10), signal transduction pathways (15) and transcription factors (21). The last group of expressed transcripts showed homology to genes of unknown function (22). To further analyze and validate our cDNA-AFLP experiments, the expression of 9 TDFs during cold acclimatzatiion was confirmed using real time RT-PCR. The results of this research show that cDNA-AFLP is a powerful technique for investigating the expression pattern of chickpea genes under low-temperature stress. Moreover, our findings will help both to elucidate the molecular basis of low-temperature effects on the chickpea genome and to identify those genes that could increase the cold tolerance of the chickpea plant.
Conflict of interest statement
Figures

References
-
- Rodríguez M, Canales E, Hidalgo OB (2005) Molecular aspects of abiotic stress in plants. Biotecnol Apl 22: 1–10.
-
- Wang WX, Vinocur B, Shoseyov O, Altman A (2001) Biotechnology of plant osmotic stress tolerance: physiological and molecular considerations. Acta Hort 560: 285–92.
-
- Heidarvand L, Maali Amiri R (2010) What happens in plant molecular responses to cold stress?. Acta Physiol Plant 32: 419–431.
-
- Chinnusamy V, Zhu J, Zhu JK (2007) Cold stress regulation of gene expression in plants. Trends Plant Sci 12: 444–451. - PubMed
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

