Transcriptional profiling of human dendritic cell populations and models--unique profiles of in vitro dendritic cells and implications on functionality and applicability
- PMID: 23341914
- PMCID: PMC3544800
- DOI: 10.1371/journal.pone.0052875
Transcriptional profiling of human dendritic cell populations and models--unique profiles of in vitro dendritic cells and implications on functionality and applicability
Abstract
Background: Dendritic cells (DCs) comprise heterogeneous populations of cells, which act as central orchestrators of the immune response. Applicability of primary DCs is restricted due to their scarcity and therefore DC models are commonly employed in DC-based immunotherapy strategies and in vitro tests assessing DC function. However, the interrelationship between the individual in vitro DC models and their relative resemblance to specific primary DC populations remain elusive.
Objective: To describe and assess functionality and applicability of the available in vitro DC models by using a genome-wide transcriptional approach.
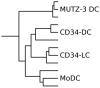
Methods: Transcriptional profiling was performed with four commonly used in vitro DC models (MUTZ-3-DCs, monocyte-derived DCs, CD34-derived DCs and Langerhans cells (LCs)) and nine primary DC populations (dermal DCs, LCs, blood and tonsillar CD123(+), CD1c(+) and CD141(+) DCs, and blood CD16(+) DCs).
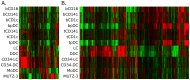
Results: Principal Component Analysis showed that transcriptional profiles of each in vitro DC model most closely resembled CD1c(+) and CD141(+) tonsillar myeloid DCs (mDCs) among primary DC populations. Thus, additional differentiation factors may be required to generate model DCs that more closely resemble other primary DC populations. Also, no model DC stood out in terms of primary DC resemblance. Nevertheless, hierarchical clustering showed clusters of differentially expressed genes among individual DC models as well as primary DC populations. Furthermore, model DCs were shown to differentially express immunologically relevant transcripts and transcriptional signatures identified for each model DC included several immune-associated transcripts.
Conclusion: The unique transcriptional profiles of in vitro DC models suggest distinct functionality in immune applications. The presented results will aid in the selection of an appropriate DC model for in vitro assays and assist development of DC-based immunotherapy.
Conflict of interest statement
Figures


Similar articles
-
Gene family clustering identifies functionally associated subsets of human in vivo blood and tonsillar dendritic cells.J Immunol. 2005 Oct 15;175(8):4839-46. doi: 10.4049/jimmunol.175.8.4839. J Immunol. 2005. PMID: 16210585
-
Phenotypic and Transcriptomic Analysis of Peripheral Blood Plasmacytoid and Conventional Dendritic Cells in Early Drug Naïve Rheumatoid Arthritis.Front Immunol. 2018 May 9;9:755. doi: 10.3389/fimmu.2018.00755. eCollection 2018. Front Immunol. 2018. PMID: 29867920 Free PMC article.
-
Profiling dendritic cell subsets in head and neck squamous cell tonsillar cancer and benign tonsils.Sci Rep. 2018 May 23;8(1):8030. doi: 10.1038/s41598-018-26193-y. Sci Rep. 2018. PMID: 29795118 Free PMC article.
-
Complexity of dendritic cell subsets and their function in the host immune system.Immunology. 2011 Aug;133(4):409-19. doi: 10.1111/j.1365-2567.2011.03457.x. Epub 2011 Jun 1. Immunology. 2011. PMID: 21627652 Free PMC article. Review.
-
The development and function of dendritic cell populations and their regulation by miRNAs.Protein Cell. 2017 Jul;8(7):501-513. doi: 10.1007/s13238-017-0398-2. Epub 2017 Mar 31. Protein Cell. 2017. PMID: 28364278 Free PMC article. Review.
Cited by
-
Re-Emergence of Dendritic Cell Vaccines for Cancer Treatment.Trends Cancer. 2018 Feb;4(2):119-137. doi: 10.1016/j.trecan.2017.12.007. Trends Cancer. 2018. PMID: 29458962 Free PMC article. Review.
-
An adjuvant-containing cDC1-targeted recombinant fusion vaccine conveys strong protection against murine melanoma growth and metastasis.Oncoimmunology. 2022 Aug 24;11(1):2115618. doi: 10.1080/2162402X.2022.2115618. eCollection 2022. Oncoimmunology. 2022. PMID: 36046810 Free PMC article.
-
NKL Homeobox Gene VENTX Is Part of a Regulatory Network in Human Conventional Dendritic Cells.Int J Mol Sci. 2021 May 31;22(11):5902. doi: 10.3390/ijms22115902. Int J Mol Sci. 2021. PMID: 34072771 Free PMC article.
-
GM-CSF Inhibits c-Kit and SCF Expression by Bone Marrow-Derived Dendritic Cells.Front Immunol. 2017 Feb 16;8:147. doi: 10.3389/fimmu.2017.00147. eCollection 2017. Front Immunol. 2017. PMID: 28261209 Free PMC article.
-
Distinctive Responses in an In Vitro Human Dendritic Cell-Based System upon Stimulation with Different Influenza Vaccine Formulations.Vaccines (Basel). 2017 Aug 9;5(3):21. doi: 10.3390/vaccines5030021. Vaccines (Basel). 2017. PMID: 28792466 Free PMC article.
References
-
- Steinman RM, Banchereau J (2007) Taking dendritic cells into medicine. Nature 449: 419–426. - PubMed
-
- Aeby P, Python F, Goebel C (2007) Skin sensitization: understanding the in vivo situation for the development of reliable in vitro test approaches. ALTEX 24 (Spec No) 3–5. - PubMed
-
- Caux C, Dezutter-Dambuyant C, Schmitt D, Banchereau J (1992) GM-CSF and TNF-alpha cooperate in the generation of dendritic Langerhans cells. Nature 360: 258–261. - PubMed
-
- Rosenzwajg M, Canque B, Gluckman JC (1996) Human dendritic cell differentiation pathway from CD34+ hematopoietic precursor cells. Blood 87: 535–544. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

