Dialysis purification of integrase-DNA complexes provides high-resolution atomic force microscopy images: dimeric recombinant HIV-1 integrase binding and specific looping on DNA
- PMID: 23341952
- PMCID: PMC3544922
- DOI: 10.1371/journal.pone.0053572
Dialysis purification of integrase-DNA complexes provides high-resolution atomic force microscopy images: dimeric recombinant HIV-1 integrase binding and specific looping on DNA
Abstract
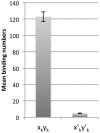
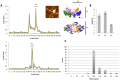
It remains difficult to obtain high-resolution atomic force microscopy images of HIV-1 integrase bound to DNA in a dimeric or tetrameric fashion. We therefore constructed specific target DNAs to assess HIV-1 integrase binding and purified the complex by dialysis prior to analysis. Our resulting atomic force microscopy analyses indicated precise size of binding human immunodeficiency virus type 1 (HIV-1) recombinant integrase in a tetrameric manner, inducing formation of a loop-like or figure-eight-like secondary structure in the target DNA. Our findings regarding the target DNA secondary structure provide new insights into the intermediate states of retroviral integration.
Conflict of interest statement
Figures


Similar articles
-
Nucleoprotein intermediates in HIV-1 DNA integration visualized by atomic force microscopy.J Mol Biol. 2010 Jun 11;399(3):491-500. doi: 10.1016/j.jmb.2010.04.026. Epub 2010 Apr 21. J Mol Biol. 2010. PMID: 20416324 Free PMC article.
-
Human immunodeficiency virus type 1 integrase: arrangement of protein domains in active cDNA complexes.EMBO J. 2001 Jul 2;20(13):3565-76. doi: 10.1093/emboj/20.13.3565. EMBO J. 2001. PMID: 11432843 Free PMC article.
-
Characterization of recombinant integrase of human immunodeficiency virus type 1 (isolate Bru).Biochemistry (Mosc). 2003 Sep;68(9):988-93. doi: 10.1023/a:1026060512380. Biochemistry (Mosc). 2003. PMID: 14606941
-
Activity of recombinant HIV-1 integrase on mini-HIV DNA.Nucleic Acids Res. 1999 May 15;27(10):2202-10. doi: 10.1093/nar/27.10.2202. Nucleic Acids Res. 1999. PMID: 10219094 Free PMC article.
-
Applications of Atomic Force Microscopy in HIV-1 Research.Viruses. 2022 Mar 21;14(3):648. doi: 10.3390/v14030648. Viruses. 2022. PMID: 35337055 Free PMC article. Review.
Cited by
-
HIV-1 Integrase Binds the Viral RNA Genome and Is Essential during Virion Morphogenesis.Cell. 2016 Aug 25;166(5):1257-1268.e12. doi: 10.1016/j.cell.2016.07.044. Cell. 2016. PMID: 27565348 Free PMC article.
-
HIV-1 integrase multimerization as a therapeutic target.Curr Top Microbiol Immunol. 2015;389:93-119. doi: 10.1007/82_2015_439. Curr Top Microbiol Immunol. 2015. PMID: 25778682 Free PMC article. Review.
-
Going beyond Integration: The Emerging Role of HIV-1 Integrase in Virion Morphogenesis.Viruses. 2020 Sep 9;12(9):1005. doi: 10.3390/v12091005. Viruses. 2020. PMID: 32916894 Free PMC article. Review.
References
-
- Liano M, Saenz DT, Meehan A, Wongthida P, Peretz M, et al. (2006) An essential role for LEDGF/p75 in HIV integration. Science 314: 461–464. - PubMed
-
- Cherepanov P, Maertens G, Proost P, Devreese B, Van Beeumen J, et al. (2003) HIV-1 integrase forms stable tetramers and associates with LEDGF/p75 protein in human cells. J Biol Chem 278: 372–381. - PubMed
-
- Christ F, Voet A, Marchand A, Nicolet S, Desimmie BA, et al. (2010) Rational design of small-molecule inhibitors of the LEDGF/p75-integrase interaction and HIV replication. Nat Chem Biol 6: 442–448. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

