N-acetylglucosamine kinase, HXK1 is involved in morphogenetic transition and metabolic gene expression in Candida albicans
- PMID: 23341961
- PMCID: PMC3544915
- DOI: 10.1371/journal.pone.0053638
N-acetylglucosamine kinase, HXK1 is involved in morphogenetic transition and metabolic gene expression in Candida albicans
Abstract
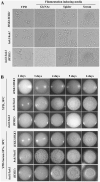
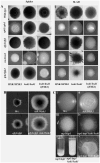
Candida albicans, a common fungal pathogen which diverged from the baker's yeast Saccharomyces cerevisiae has the unique ability to utilise N-acetylglucosamine, an amino sugar and exhibits phenotypic differences. It has acquired intricate regulatory mechanisms at different levels in accordance with its life style. N-acetylglucosamine kinase, a component of the N-acetylglucosamine catabolic cascade is an understudied gene since Saccharomyces cerevisiae lacks it. We report HXK1 to act as both positive and negative regulator of transcription of genes involved in maintaining cellular homeostasis. It is involved in repression of hyphal specific genes in addition to metabolic genes. Its regulation of filamentation and GlcNAc metabolism is independent of the known classical regulators like EFG1, CPH1, RAS1, TPK2 or TUP1. Moreover, Hxk1-GFP is localised to cytoplasm, nucleus and mitochondria in a condition specific manner. By employing two-step affinity purification, we report the interaction of HXK1 with SIR2 under filamentation inducing conditions. Our work highlights a novel regulatory mechanism involved in filamentation repression and attempts to decipher the GlcNAc catabolic regulatory cascade in eukaryotes.
Conflict of interest statement
Figures







Similar articles
-
N-acetylglucosamine kinase, HXK1 contributes to white-opaque morphological transition in Candida albicans.Biochem Biophys Res Commun. 2014 Feb 28;445(1):138-44. doi: 10.1016/j.bbrc.2014.01.123. Epub 2014 Jan 31. Biochem Biophys Res Commun. 2014. PMID: 24491547
-
Role of the N-acetylglucosamine kinase (Hxk1) in the regulation of white-gray-opaque tristable phenotypic transitions in C. albicans.Fungal Genet Biol. 2016 Jul;92:26-32. doi: 10.1016/j.fgb.2016.05.001. Epub 2016 May 3. Fungal Genet Biol. 2016. PMID: 27153757
-
N-acetylglucosamine kinase, Hxk1 is a multifaceted metabolic enzyme in model pathogenic yeast Candida albicans.Microbiol Res. 2022 Oct;263:127146. doi: 10.1016/j.micres.2022.127146. Epub 2022 Jul 26. Microbiol Res. 2022. PMID: 35940108 Review.
-
Molecular Characterization of the N-Acetylglucosamine Catabolic Genes in Candida africana, a Natural N-Acetylglucosamine Kinase (HXK1) Mutant.PLoS One. 2016 Jan 25;11(1):e0147902. doi: 10.1371/journal.pone.0147902. eCollection 2016. PLoS One. 2016. PMID: 26808192 Free PMC article.
-
Co-regulation of pathogenesis with dimorphism and phenotypic switching in Candida albicans, a commensal and a pathogen.Int J Med Microbiol. 2002 Oct;292(5-6):299-311. doi: 10.1078/1438-4221-00215. Int J Med Microbiol. 2002. PMID: 12452278 Review.
Cited by
-
Hexokinase and Glucokinases Are Essential for Fitness and Virulence in the Pathogenic Yeast Candida albicans.Front Microbiol. 2019 Feb 25;10:327. doi: 10.3389/fmicb.2019.00327. eCollection 2019. Front Microbiol. 2019. PMID: 30858840 Free PMC article.
-
The N-Acetylglucosamine Kinase from Yarrowia lipolytica Is a Moonlighting Protein.Int J Mol Sci. 2021 Dec 3;22(23):13109. doi: 10.3390/ijms222313109. Int J Mol Sci. 2021. PMID: 34884915 Free PMC article.
-
The Absence of N-Acetyl-D-glucosamine Causes Attenuation of Virulence of Candida albicans upon Interaction with Vaginal Epithelial Cells In Vitro.Biomed Res Int. 2015;2015:398045. doi: 10.1155/2015/398045. Epub 2015 Aug 20. Biomed Res Int. 2015. PMID: 26366412 Free PMC article.
-
The roles of ING5 expression in ovarian carcinogenesis and subsequent progression: a target of gene therapy.Oncotarget. 2017 Oct 19;8(61):103449-103464. doi: 10.18632/oncotarget.21968. eCollection 2017 Nov 28. Oncotarget. 2017. PMID: 29262575 Free PMC article.
-
Multilocus sequence typing analysis of Candida africana from vulvovaginal candidiasis.BMC Infect Dis. 2019 May 22;19(1):461. doi: 10.1186/s12879-019-4071-7. BMC Infect Dis. 2019. PMID: 31117966 Free PMC article.
References
-
- Biswas M, Singh B, Datta A (1979) Induction of N-acetylmannosamine catabolic pathway in yeast. Biochim Biophys Acta 585: 535–542. - PubMed
-
- Shepherd MG, Yin CY, Ram SP, Sullivan PA (1980) Germ tube induction in Candida albicans . Can J Microbiol 26: 21–26. - PubMed
-
- Sudbery P, Gow N, Berman J (2004) The distinct morphogenic states of Candida albicans. . Trends Microbiol 12: 317. - PubMed
-
- Liu H (2001) Transcriptional control of dimorphism in Candida albicans. . Curr Opin Microbiol 4: 278–235. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

