Biased diversity metrics revealed by bacterial 16S pyrotags derived from different primer sets
- PMID: 23341963
- PMCID: PMC3544912
- DOI: 10.1371/journal.pone.0053649
Biased diversity metrics revealed by bacterial 16S pyrotags derived from different primer sets
Abstract
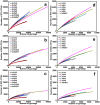
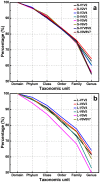
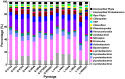
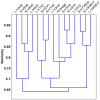
In recent years, PCR-based pyrosequencing of 16S rRNA genes has continuously increased our understanding of complex microbial communities in various environments of the Earth. However, there is always concern on the potential biases of diversity determination using different 16S rRNA gene primer sets and covered regions. Here, we first report how bacterial 16S rRNA gene pyrotags derived from a series of different primer sets resulted in the biased diversity metrics. In total, 14 types of pyrotags were obtained from two-end pyrosequencing of 7 amplicon pools generated by 7 primer sets paired by 1 of 4 forward primers (V1F, V3F, V5F, and V7F) and 1 of 4 reverse primers (V2R, V4R, V6R, and V9R), respectively. The results revealed that: i) the activated sludge exhibited a large bacterial diversity that represented a broad range of bacterial populations and served as a good sample in this methodology research; ii) diversity metrics highly depended on the selected primer sets and covered regions; iii) paired pyrotags obtained from two-end pyrosequencing of each short amplicon displayed different diversity metrics; iv) relative abundance analysis indicated the sequencing depth affected the determination of rare bacteria but not abundant bacteria; v) the primer set of V1F and V2R significantly underestimated the diversity of activated sludge; and vi) the primer set of V3F and V4R was highly recommended for future studies due to its advantages over other primer sets. All of these findings highlight the significance of this methodology research and offer a valuable reference for peer researchers working on microbial diversity determination.
Conflict of interest statement
Figures



References
-
- Wagner M, Noguera DR, Juretschko S, Rath G, Koops HP, et al. (1998) Combining fluorescent in situ hybridization (FISH) with cultivation and mathematical modeling to study population structure and function of ammonia-oxidizing bacteria in activated sludge. Water Sci Technol 37: 441–449.
-
- He ZL, Van Nostrand JD, Zhou JZ (2012) Applications of functional gene microarrays for profiling microbial communities. Curr Opin Biotech 23: 460–466. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

