Inferring hierarchical orthologous groups from orthologous gene pairs
- PMID: 23342000
- PMCID: PMC3544860
- DOI: 10.1371/journal.pone.0053786
Inferring hierarchical orthologous groups from orthologous gene pairs
Abstract
Hierarchical orthologous groups are defined as sets of genes that have descended from a single common ancestor within a taxonomic range of interest. Identifying such groups is useful in a wide range of contexts, including inference of gene function, study of gene evolution dynamics and comparative genomics. Hierarchical orthologous groups can be derived from reconciled gene/species trees but, this being a computationally costly procedure, many phylogenomic databases work on the basis of pairwise gene comparisons instead ("graph-based" approach). To our knowledge, there is only one published algorithm for graph-based hierarchical group inference, but both its theoretical justification and performance in practice are as of yet largely uncharacterised. We establish a formal correspondence between the orthology graph and hierarchical orthologous groups. Based on that, we devise GETHOGs ("Graph-based Efficient Technique for Hierarchical Orthologous Groups"), a novel algorithm to infer hierarchical groups directly from the orthology graph, thus without needing gene tree inference nor gene/species tree reconciliation. GETHOGs is shown to correctly reconstruct hierarchical orthologous groups when applied to perfect input, and several extensions with stringency parameters are provided to deal with imperfect input data. We demonstrate its competitiveness using both simulated and empirical data. GETHOGs is implemented as a part of the freely-available OMA standalone package (http://omabrowser.org/standalone). Furthermore, hierarchical groups inferred by GETHOGs ("OMA HOGs") on >1,000 genomes can be interactively queried via the OMA browser (http://omabrowser.org).
Conflict of interest statement
Figures
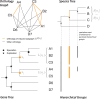
 are drawn in orange. By definition, these groups correspond to the sets of leaves attached to the speciation nodes of the gene tree coloured in orange.
are drawn in orange. By definition, these groups correspond to the sets of leaves attached to the speciation nodes of the gene tree coloured in orange.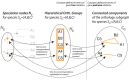
 (right). Lemma 1 establishes the one-to-one correspondence between
(right). Lemma 1 establishes the one-to-one correspondence between  and
and  (which we prove by viewing it as composition of the one-to-one correspondences
(which we prove by viewing it as composition of the one-to-one correspondences  and
and  ).
).
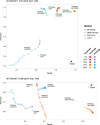
 parameter values for GETHOGs and bootstrap value for COCO-CL.
parameter values for GETHOGs and bootstrap value for COCO-CL.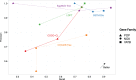
 and bootstrap
and bootstrap ). The points for EggNOG and OrthoDB are from the original analysis (Reference [9],table 2).
). The points for EggNOG and OrthoDB are from the original analysis (Reference [9],table 2).References
-
- Fitch WM (1970) Distinguishing homologous from analogous proteins. Syst Zool 19: 99–113. - PubMed
-
- Altenhoff AM, Dessimoz C (2012) Inferring orthology and paralogy. In: Anisimova M, editor, Evolutionary Genomics, Clifton, NJ, USA: Methods in Molecular Biology. 259–279. - PubMed
-
- Wall DP, Fraser HB, Hirsh AE (2003) Detecting putative orthologs. Bioinformatics 19: 1710–1711. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

