Expression and functional studies on the noncoding RNA, PRINS
- PMID: 23344029
- PMCID: PMC3565259
- DOI: 10.3390/ijms14010205
Expression and functional studies on the noncoding RNA, PRINS
Abstract
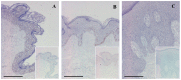
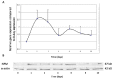
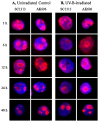
PRINS, a noncoding RNA identified earlier by our research group, contributes to psoriasis susceptibility and cellular stress response. We have now studied the cellular and histological distribution of PRINS by using in situ hybridization and demonstrated variable expressions in different human tissues and a consistent staining pattern in epidermal keratinocytes and in vitro cultured keratinocytes. To identify the cellular function(s) of PRINS, we searched for a direct interacting partner(s) of this stress-induced molecule. In HaCaT and NHEK cell lysates, the protein proved to be nucleophosmin (NPM) protein as a potential physical interactor with PRINS. Immunohistochemical experiments revealed an elevated expression of NPM in the dividing cells of the basal layers of psoriatic involved skin samples as compared with healthy and psoriatic uninvolved samples. Others have previously shown that NPM is a ubiquitously expressed nucleolar phosphoprotein which shuttles to the nucleoplasm after UV-B irradiation in fibroblasts and cancer cells. We detected a similar translocation of NPM in UV-B-irradiated cultured keratinocytes. The gene-specific silencing of PRINS resulted in the retention of NPM in the nucleolus of UV-B-irradiated keratinocytes; suggesting that PRINS may play a role in the NPM-mediated cellular stress response in the skin.
Figures





References
-
- Lebwohl M. Psoriasis. Lancet. 2003;361:1197–1204. - PubMed
-
- Bowcock A.M., Krueger J.G. Getting under the skin: The immunogenetics of psoriasis. Nat. Rev. Immunol. 2005;5:699–711. - PubMed
-
- Duffy D.L., Spelman L.S., Martin N.G. Psoriasis in Australian twins. J. Am. Acad. Dermatol. 1993;29:428–434. - PubMed
-
- Bos J.D., De Rie M.A., Teunissen M.B., Piskin G. Psoriasis: Ddysregulation of innate immunity. Br. J. Dermatol. 2005;152:1098–1107. - PubMed
-
- Bos J.D., Hulsebosch H.J., Krieg S.R., Bakker P.M., Cormane R.H. Immunocompetent cells in psoriasis. In situ immunophenotyping by monoclonal antibodies. Arch. Dermatol. Res. 1983;275:181–189. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

