Quantitative profiling of DNA damage and apoptotic pathways in UV damaged cells using PTMScan Direct
- PMID: 23344034
- PMCID: PMC3565264
- DOI: 10.3390/ijms14010286
Quantitative profiling of DNA damage and apoptotic pathways in UV damaged cells using PTMScan Direct
Abstract
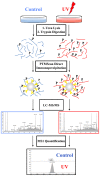
Traditional methods for analysis of peptides using liquid chromatography and tandem mass spectrometry (LC-MS/MS) lack the specificity to comprehensively monitor specific biological processes due to the inherent duty cycle limitations of the MS instrument and the stochastic nature of the analytical platform. PTMScan Direct is a novel, antibody-based method that allows quantitative LC-MS/MS profiling of specific peptides from proteins that reside in the same signaling pathway. New PTMScan Direct reagents have been produced that target peptides from proteins involved in DNA Damage/Cell Cycle and Apoptosis/Autophagy pathways. Together, the reagents provide access to 438 sites on 237 proteins in these signaling cascades. These reagents have been used to profile the response to UV damage of DNA in human cell lines. UV damage was shown to activate canonical DNA damage response pathways through ATM/ATR-dependent signaling, stress response pathways and induce the initiation of apoptosis, as assessed by an increase in the abundance of peptides corresponding to cleaved, activated caspases. These data demonstrate the utility of PTMScan Direct as a multiplexed assay for profiling specific cellular responses to various stimuli, such as UV damage of DNA.
Figures



Similar articles
-
PTMScan direct: identification and quantification of peptides from critical signaling proteins by immunoaffinity enrichment coupled with LC-MS/MS.Mol Cell Proteomics. 2012 May;11(5):187-201. doi: 10.1074/mcp.M111.015883. Epub 2012 Feb 9. Mol Cell Proteomics. 2012. PMID: 22322096 Free PMC article.
-
Profiling of UV-induced ATM/ATR signaling pathways.Proc Natl Acad Sci U S A. 2007 Dec 11;104(50):19855-60. doi: 10.1073/pnas.0707579104. Epub 2007 Dec 6. Proc Natl Acad Sci U S A. 2007. PMID: 18077418 Free PMC article.
-
Isobaric Tandem Mass Tag Multiplexed Post-Translational Modification Quantitation of Biopharmaceuticals by Targeted High-Resolution Mass Spectrometry.Anal Chem. 2020 Jul 21;92(14):9682-9690. doi: 10.1021/acs.analchem.0c00999. Epub 2020 Jun 30. Anal Chem. 2020. PMID: 32559367
-
Mass spectrometry-based proteomic analysis of the DNA damage response.Front Biosci (Landmark Ed). 2018 Jan 1;23(4):597-613. doi: 10.2741/4607. Front Biosci (Landmark Ed). 2018. PMID: 28930563 Review.
-
How DNA lesions are turned into powerful killing structures: insights from UV-induced apoptosis.Mutat Res. 2009 Mar-Jun;681(2-3):197-208. doi: 10.1016/j.mrrev.2008.09.001. Epub 2008 Sep 19. Mutat Res. 2009. PMID: 18845270 Review.
Cited by
-
Complementary PTM Profiling of Drug Response in Human Gastric Carcinoma by Immunoaffinity and IMAC Methods with Total Proteome Analysis.Proteomes. 2015 Aug 7;3(3):160-183. doi: 10.3390/proteomes3030160. Proteomes. 2015. PMID: 28248267 Free PMC article.
-
Caspases and their substrates.Cell Death Differ. 2017 Aug;24(8):1380-1389. doi: 10.1038/cdd.2017.44. Epub 2017 May 12. Cell Death Differ. 2017. PMID: 28498362 Free PMC article. Review.
-
Systems-wide analysis of K-Ras, Cdc42, and PAK4 signaling by quantitative phosphoproteomics.Mol Cell Proteomics. 2013 Aug;12(8):2070-80. doi: 10.1074/mcp.M112.027052. Epub 2013 Apr 22. Mol Cell Proteomics. 2013. PMID: 23608596 Free PMC article.
-
Involvement of Akt/mTOR in the Neurotoxicity of Rotenone-Induced Parkinson's Disease Models.Int J Environ Res Public Health. 2019 Oct 10;16(20):3811. doi: 10.3390/ijerph16203811. Int J Environ Res Public Health. 2019. PMID: 31658620 Free PMC article.
-
Proteomics of protein post-translational modifications implicated in neurodegeneration.Transl Neurodegener. 2014 Oct 30;3(1):23. doi: 10.1186/2047-9158-3-23. eCollection 2014. Transl Neurodegener. 2014. PMID: 25671099 Free PMC article. Review.
References
-
- Abraham R.T. Cell cycle checkpoint signaling through the ATM and ATR kinases. Genes Dev. 2001;15:2177–2196. - PubMed
-
- Hartwell L.H., Weinert T.A. Checkpoints: Controls that ensure the order of cell cycle events. Science. 1989;246:629–634. - PubMed
-
- Zhou B.B., Elledge S.J. The DNA damage response: putting checkpoints in perspective. Nature. 2000;408:433–439. - PubMed
-
- Batista L.F., Kaina B., Meneghini R., Menck C.F. How DNA lesions are turned into powerful killing structures: insights from UV-induced apoptosis. Mutat. Res. 2009;681:197–208. - PubMed
-
- Izumaru S., Arima N., Toyozumi Y., Kato S., Morimatsu M., Nakashima T. Down-regulation of p21Waf-1 protein facilitates IR- and UV-induced apoptosis in human squamous carcinoma cells. Int. J. Oncol. 2004;24:1245–1255. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

