Genomic island genes in a coastal marine Synechococcus strain confer enhanced tolerance to copper and oxidative stress
- PMID: 23344240
- PMCID: PMC3660668
- DOI: 10.1038/ismej.2012.175
Genomic island genes in a coastal marine Synechococcus strain confer enhanced tolerance to copper and oxidative stress
Abstract
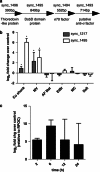
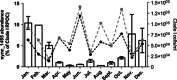
Highly variable regions called genomic islands are found in the genomes of marine picocyanobacteria, and have been predicted to be involved in niche adaptation and the ecological success of these microbes. These picocyanobacteria are typically highly sensitive to copper stress and thus, increased copper tolerance could confer a selective advantage under some conditions seen in the marine environment. Through targeted gene inactivation of genomic island genes that were known to be upregulated in response to copper stress in Synechococcus sp. strain CC9311, we found two genes (sync_1495 and sync_1217) conferred tolerance to both methyl viologen and copper stress in culture. The prevalence of one gene, sync_1495, was then investigated in natural samples, and had a predictable temporal variability in abundance at a coastal monitoring site with higher abundance in winter months. Together, this shows that genomic island genes can confer an adaptive advantage to specific stresses in marine Synechococcus, and may help structure their population diversity.
Figures
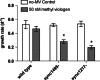


References
-
- Avrani S, Wurtzel O, Sharon I, Sorek R, Lindell D. Genomic island variability facilitates Prochlorococcus-virus coexistence. Nature. 2011;474:604–608. - PubMed
-
- Baron M, Arellano JB, Gorge JL. Copper and photosystem-II - A controversial relationship. Physiol Plant. 1995;94:174–180.
-
- Blake AC, Chadwick DB, Zirino A, Rivera-Duarte I. Spatial and temporal variations in copper speciation in San Diego Bay. Estuaries. 2004;27:437–447.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

